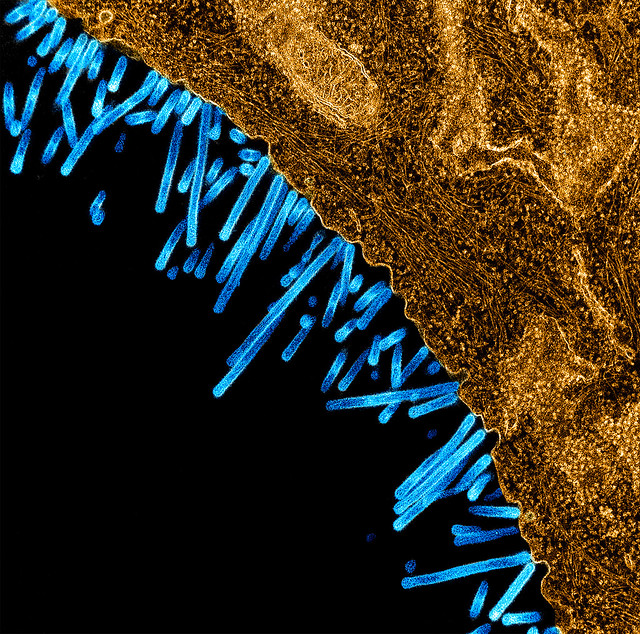
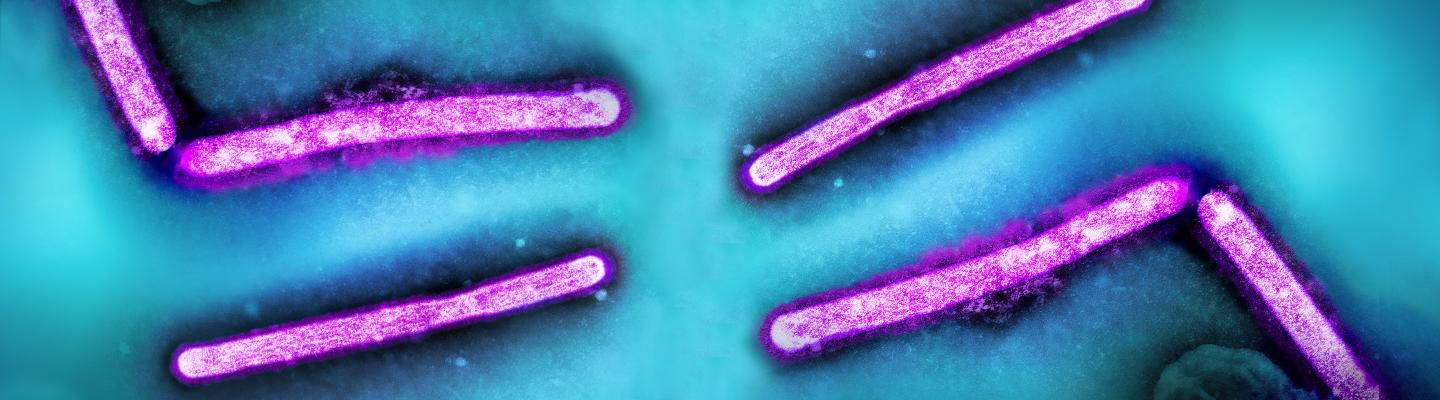
NIAID has a longstanding commitment to conducting and supporting the basic research necessary to understand how influenza strains emerge, evolve, infect and cause disease (called pathogenesis) in animals and humans. Results from this research are used to inform the design of new and improved influenza vaccines, diagnostics and antiviral drugs to treat flu infection.
Influenza is challenging for scientists to study because there are hundreds of strains that are classified into four main categories: A through D, though D is not known to infect people. Influenza A virus is the group that most commonly causes illness in humans and is the source of all of the major influenza pandemics in modern history. This type can drift and shift through birds and animals, meaning it emerges with rearranged surface proteins that create different strains of the virus.
The surface proteins that combine in different ways to create an assortment of influenza virus type A strains are called hemagglutinin (HA) and neuraminidase (NA). Hemagglutinin enables the flu virus to enter a human cell and initiate infection; neuraminidase allows newly formed flu viruses to exit the host cell and multiply throughout the body. There are 18 types of HA and 11 types of NA, leaving the possibility for dozens of different subtypes of influenza A viruses (such as H1N1, H3N2, H5N8, and H7N9) and strains (such as 1918 H1N1 influenza and 2009 H1N1 flu).
Some of the specific questions about influenza that basic science researchers explore include how strains of viruses differ from each other by gene structure; how viruses can defeat the immune system to cause disease; how some viruses can transmit from person to person; and how some treatments and vaccines effectively prevent or minimize infection.
Specific examples of NIAID influenza basic research include:
- Trying to determine if hyperimmune plasma – the liquid component of blood that contains antibodies to help to clear the virus from the body – is an effective treatment against influenza, specifically for patients at high risk for developing severe disease. Similar work is exploring the use of hyperimmune immunoglobulin (purified antibodies).
- Developing human and animal models to observe how influenza virus enters a host – and documenting the specific processes that occur and the structures within the virus and the host that contribute to infection. Then, similar to reading a road map, scientists try and introduce biological “detours” to both avoid a route to infection (preventive vaccination) and improve recovery time from infection (therapeutic treatment).
- Studying the relationship between influenza and co-infection with bacteria, such as Staphylococcus aureus or Streptococcus pneumoniae. Influenza can lead to viral and bacterial pneumonia (most deaths attributed to influenza virus are caused by pneumonia). The studies use an animal model to assess damage to the lungs and the amount of oxygen reaching the bloodstream. This approach will help researchers assess the effectiveness of new vaccines and therapeutics at stopping disease.
- Exposing healthy volunteers under carefully controlled and monitored conditions to influenza A viruses. These types of challenge studies provide critical information as to how the flu develops and persists and how humans fight infection. This information is key to establishing more rapid, cost-effective clinical trials for new influenza medicines or for determining the efficacy of candidate vaccines for preventing seasonal or pandemic influenza.
- Characterizing historic influenza A viruses to compare and understand their genetic traits to learn why some influenza virus strains of the same subtype are more severe than others. For example, 1918 H1N1 flu killed an estimated 675,000 people in the United States and was much more severe than the 2009 H1N1 influenza. Both of these virus strains are considered pandemic because they circulated around the world. After successfully sequencing the 1918 H1N1 virus in 2005, scientists are now working on the same project for influenza A viruses that circulated in the decade before and after 1918. They are hoping the comparisons will help them understand the biological scenario that allowed the deadly 1918 strain to emerge and what has happened to that virus since 1918. One goal is to help global leaders plan for the next H1N1 subtype that could cause severe disease in people and become a pandemic.
- Examining common influenza vaccination practices in an effort to develop more effective vaccines. For example, one study is exploring whether suppressing neuraminidase could have a more significant role in establishing immunity; most influenza vaccines focus on limiting concentrations of hemagglutinin. Another study is examining influenza virus epitopes – the precise locations on the HA protein where antibodies bind to prevent infection. Little is known about relationships between epitope display and vaccine effectiveness, though researchers are using specialized electron microscopy and computational analyses to build three-dimensional models to predict how and where antibodies bind to epitopes. Scientists believe that a better understanding of the molecular architecture of epitopes will help them develop more effective vaccines.
- Understanding why influenza replication is higher in some people to develop strategies to prevent disease and viral spread. A March 2016 study found that estrogen provides women, but not men, extra protection from influenza. Researchers showed how estrogen affects the ability of the influenza virus to replicate in nasal cells and why the antiviral effect is more robust in women. The results suggest that pre-menopausal or post-menopausal women who do not produce estrogen could benefit from hormone replacement during influenza season.
Scientific Advances
Influenza A Viruses Adapt Shape in Response to Environmental Pressures
February 10, 2025Influenza A virus particles strategically adapt their shape – to become either spheres or larger filaments – to favor their ability to infect cells depending on environmental conditions, a new NIAID study published in Nature Microbiology reveals. This previously unrecognized response could help explain how influenza A and other viruses persist in populations, evade immune responses, and acquire…
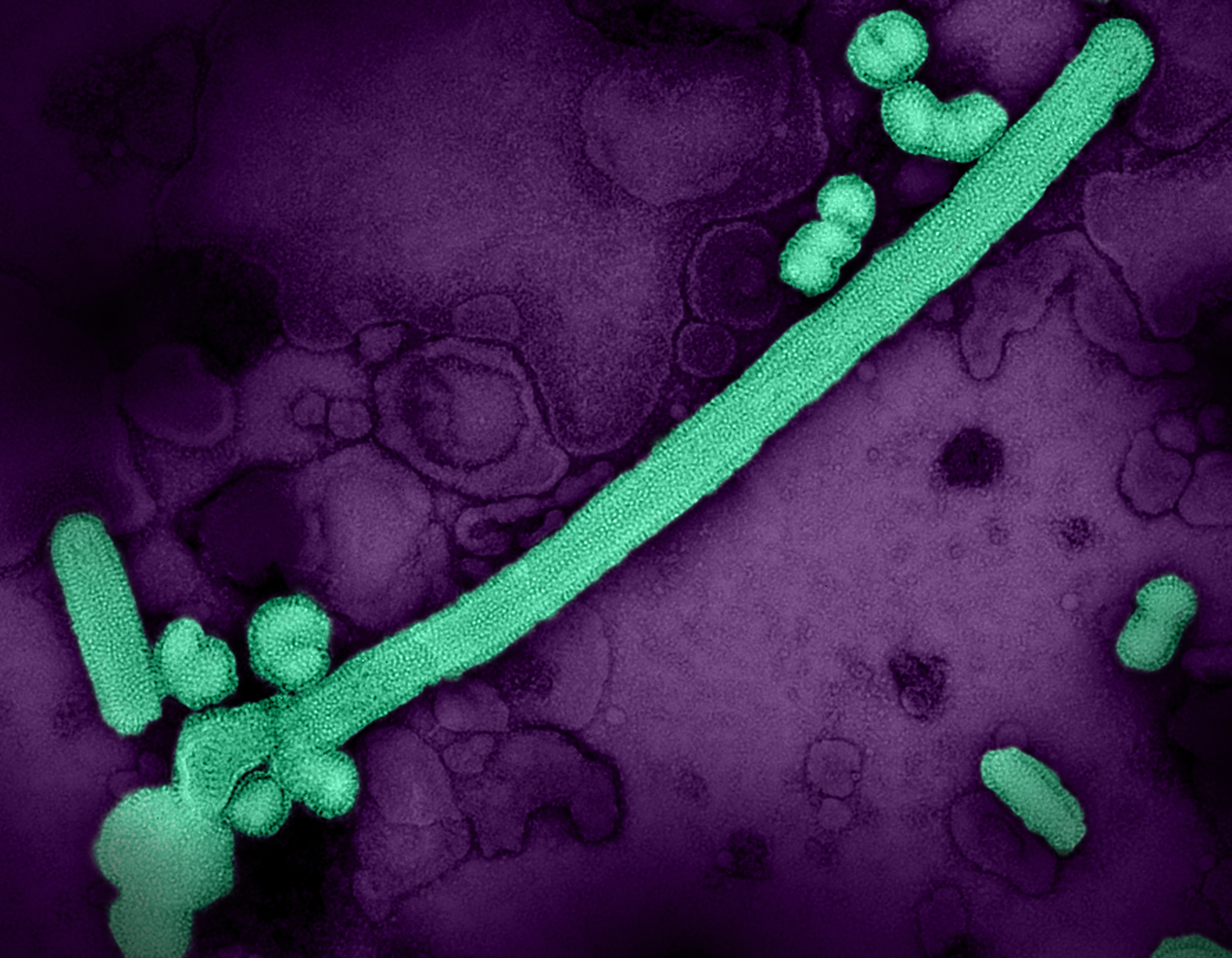
Single Mutation in H5N1 Influenza Surface Protein Could Enable Easier Human Infection
December 6, 2024A single modification in the protein found on the surface of the highly pathogenic avian influenza (HPAI) H5N1 influenza virus currently circulating in U.S. dairy cows could allow for easier transmission among humans, according to new research funded by the National Institutes of Health (NIH) and published today in the journal Science. The study results reinforce the need for continued, vigilant…
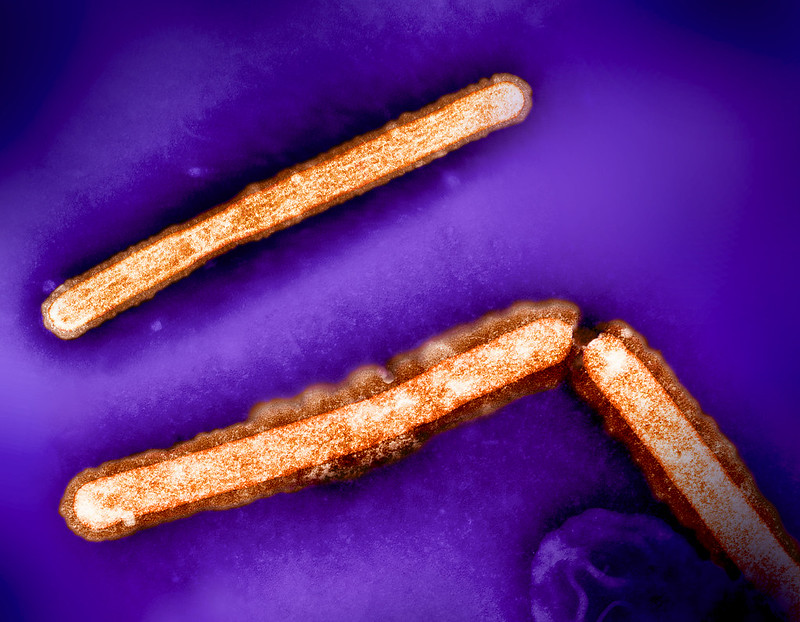
Bovine H5N1 Influenza from Infected Worker Transmissible and Lethal in Animal Models
October 28, 2024Bovine H5N1 influenza virus taken from eye of infected worker transmissible and lethal in animal models.
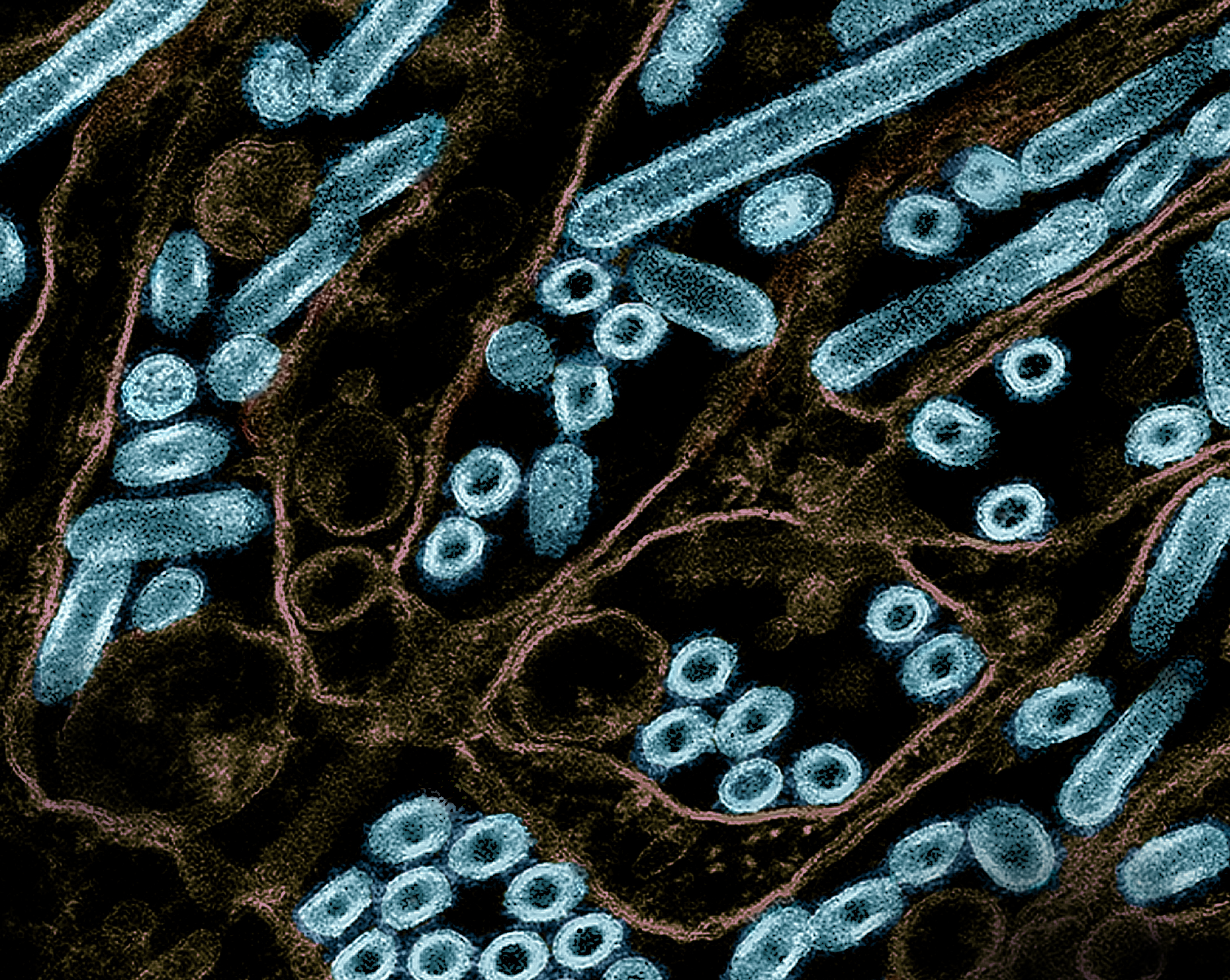
Features of H5N1 Influenza Viruses in Dairy Cows May Facilitate Infection, Transmission in Mammals
July 8, 2024A series of experiments with highly pathogenic H5N1 avian influenza (HPAI H5N1) viruses circulating in infected U.S. dairy cattle found that viruses derived from lactating dairy cattle induced severe disease in mice and ferrets when administered via intranasal inoculation.
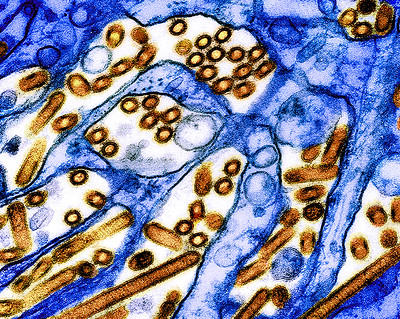
Infectious H5N1 Influenza Virus in Raw Milk Rapidly Declines with Heat Treatment
June 14, 2024The amount of infectious H5N1 influenza viruses in raw milk rapidly declined with heat treatment in laboratory research conducted by scientists at the National Institute of Allergy and Infectious Diseases (NIAID), part of the National Institutes of Health. However, small, detectable amounts of infectious virus remained in raw milk samples with high virus levels when treated at 72 degrees Celsius …

NIAID Funds New Influenza Research Network
April 14, 2021The National Institute of Allergy and Infectious Diseases (NIAID), part of the National Institutes of Health, has established a network of research sites to study the natural history, transmission and pathogenesis of influenza and provide an international research infrastructure to address influenza outbreaks.
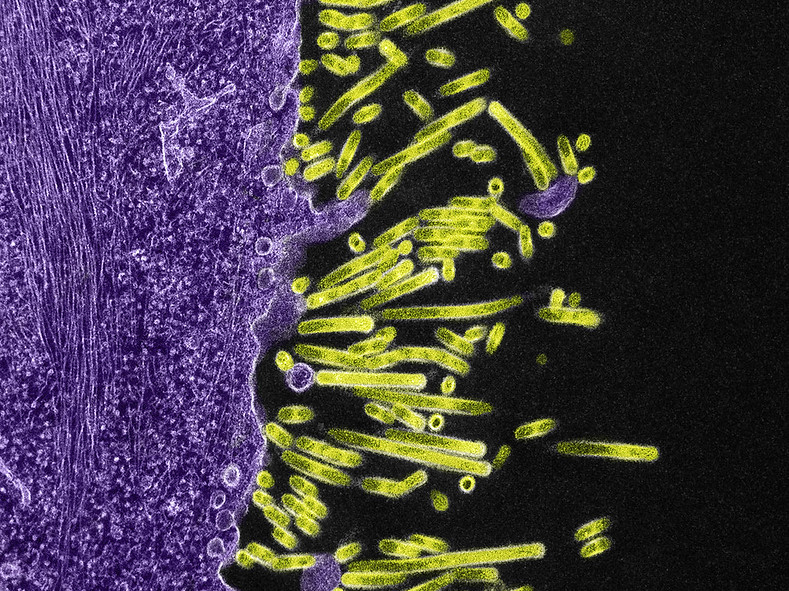
Human Antibody Reveals Hidden Vulnerability in Influenza Virus
May 16, 2019The ever-changing “head” of an influenza virus protein has an unexpected Achilles heel, report scientists funded by the National Institute of Allergy and Infectious Diseases (NIAID), one of the National Institutes of Health. The team discovered and characterized the structure of a naturally occurring human antibody that recognizes and disrupts a portion of the hemagglutinin (HA) protein that the…