Certain species of fungi are responsible for many common infections including yeast infections, ringworm, thrush, and athlete’s foot. While these diseases may not lead to serious outcomes for most healthy individuals, fungal infections can be deadly, especially for patients with weakened immune systems. One fungal pathogen, Candida auris, is an emerging healthcare-associated infection of growing public health concern. The first case of Candida auris was reported in 2009 in Japan, where it was isolated from a patient’s ear (Auris is Latin for “ear”). Outbreaks have since emerged rapidly around the globe. In the United States, C. auris infections have been increasing over the past few years, with more than 1460 cases reported in 2021. C. auris is typically found in hospitals and other healthcare settings and can cause serious bloodstream and wound infections.
Like other Candida species, C. auris is a type of yeast. However, unlike its yeasty cousins, this pathogen can colonize patients’ skin and persist for long periods of time on environmental surfaces. Another challenge is that C. auris is often resistant to one or more of the major classes of drugs that are typically used to treat fungal infections. While most C. auris infections can be treated with a class of antifungals called echinocandins, resistance to these drugs has also been reported, making some infections difficult to treat. C. auris is the only fungal pathogen identified as an ‘urgent’ threat in CDC’s Antibiotic Resistance Threat Report.
Back to basics
NIAID supports several researchers who are asking fundamental questions about the biology of C. auris. Today’s NIAID Now post features insights from NIAID-funded researchers, Jeniel Nett, M.D., Ph.D., associate professor of medicine and medical microbiology and immunology at University of Wisconsin-Madison, and Christina Cuomo, Ph.D., associate director of the Genomic Center of Infectious Diseases at the Broad Institute of Massachusetts Institute of Technology and Harvard University.
How is C. auris able to colonize the skin and persist in the environment?
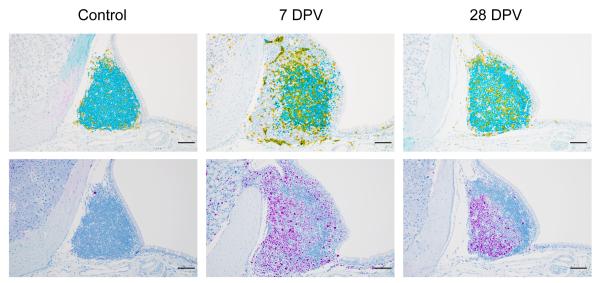
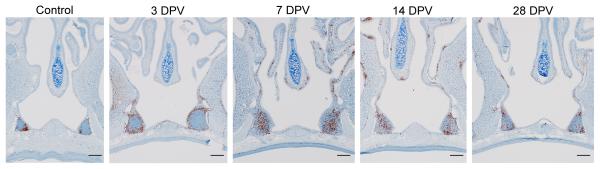
C. auris can live and grow on the skin or in the body without causing illness. However, people who are colonized with C. auris may spread the pathogen to others and are at risk of getting sick later on if they develop infections. One important question to understanding C. auris outbreaks is: how is the fungus able to colonize skin so effectively and to persist in the environment? Dr. Nett’s research group is tackling this question by studying C. auris growth in the lab using two different systems. The first is designed to mimic human sweat and skin. Nett noted, “we think that this represents skin to some degree but also when surfaces get contaminated with skin and sweat components.” The other system is pig skin. “Pigs have similar skin to humans in terms of skin thickness and some of the cell types,” Nett explained. Using these systems, Nett and colleagues have shown that C. auris is able to readily grow on skin. “It really seems to mirror what we’re seeing patients,” said Nett. They’ve found that when the fungus is grown in the synthetic sweat medium, it forms multi-layered plaques, or biofilms, both on the pig skin as well as on hard surfaces. Compared to other Candida species, the biofilms are thicker and contain more viable organisms. C. auris biofilms can also persist on surfaces without drying out for up to two weeks in the lab.

Jeniel Nett, M.D., Ph.D., associate professor of medicine and medical microbiology and immunology at University of Wisconsin-Madison
Nett’s research demonstrates the ability of the fungus to colonize skin and form persistent biofilms on environmental surfaces, which has implications for transmission in healthcare settings. “This really becomes important with reusable medical equipment that goes room to room,” Nett emphasized. The systems Nett’s group has developed to study C. auris in the lab can also inform potential strategies to remove C. auris from the skin of patients. Nett’s research has shown that while antiseptics are somewhat effective, they are not as active against C. auris when the fungus is growing in the skin environment compared to when it is growing without the skin present. In a published manuscript, Nett and colleagues demonstrated that the commonly used topical antiseptic chlorhexidine does not fully remove C. auris from the skin of patients. They also showed that by adding isopropanol, as well as some essential oils, including tea tree and lemongrass, to chlorhexidine, they were able to improve the activity of the antiseptic. Her group is still investigating what specific components of skin and sweat are triggering biofilm growth in C. auris. Understanding this could lead to better, more specific strategies to disrupt skin colonization.
How did C. auris outbreaks emerge around the world, and how has the fungus become multidrug-resistant?
Soon after it was first identified, outbreaks of C. auris arose in four distinct locations—South Asia, East Asia, Africa, and South America—nearly simultaneously. Dr. Cuomo and colleagues are using a genomics approach to better understand this phenomenon.
“One fundamental question genomics can answer is, what has been the history of the pathogen over time?” Cuomo explained. “We can take isolates from different patients, and by comparing them we can infer back in time to where they have a common connection.”

Christina Cuomo, Ph.D., associate director of the Genomic Center of Infectious Diseases at the Broad Institute of Massachusetts Institute of Technology and Harvard University
Together with colleagues at the Centers for Disease Control and Prevention, Cuomo’s group helped confirm that the different outbreaks were caused by distinct genetic groups, or ‘clades.’ As cases have continued to spread around the globe, researchers have been able to trace new C. auris isolates back to these four major clades, allowing them to understand how the different outbreaks are connected.
Expanding on this initial work, Cuomo’s group is looking more closely at the different C. auris clades and identifying key genetic differences both within and between these groups as well as among C. auris and other related Candida species. From this analysis, they have generated hypotheses about which genes in the fungus are important for contributing to disease in humans. Such studies provide important insight into the biology of C. auris and can help identify potential targets for new drugs.
Researchers are also actively trying to understand how this fungal species has evolved to become resistant to certain antifungal drugs. Combining clinical data and experimental evolution studies, Cuomo’s group has identified specific mutations, or genetic changes, contributing to resistance to the major classes of antifungal drugs, including echinocandins. Cuomo explained that a single change in one of the C. auris proteins causes the fungus to go from sensitive to resistant, which explains why patients will sometimes stop responding in the middle of treatment with echinocandins.
The genomic resources that Cuomo and her group have developed are used by public health laboratories to help assess the frequency of drug resistance in C. auris. Understanding what genetic changes are associated with drug resistance can also help inform patient treatment. “That’s the kind of information we want to be marrying to traditional diagnostics, to think about how can we best type resistance across the course of a patient’s treatment,” Cuomo noted. “We know that resistance can arise while on treatment. We’d like to detect that as soon as it emerges, and not when the patient succumbs to a very high fever or other devastating symptoms.”
From knowledge to solutions
Working on a novel pathogen is a challenging effort. Both Drs. Nett and Cuomo have forged into relatively new scientific territory, and have had to develop new tools, methods, and resources to study C. auris. However, their work has the potential to make a significant impact against this emerging disease. While the scientific questions they both are tackling are fundamental in nature, the answers are of critical importance to patient care and public health interventions.
Learn more about this research by reading recent papers from Dr. Nett, Dr. Cuomo, and colleagues:
CJ, Johnson et al. Modeling Candida auris skin colonization: Mice, swine, and humans. PLOS Pathogens. DOI: 10.1371/journal.ppat.1010730 (2022)
JM Rybak et al. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin Microbiology Infect. DOI:10.1016/j.cmi.2021.11.024 (2022).
C Johnson et al. Augmenting the Activity of Chlorhexidine for Decolonization of Candida auris from Porcine skin. J Fungi. DOI: : 10.3390/jof7100804 (2021).
J Muñoz et al. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics. DOI: : 10.1093/genetics/iyab029 (2021).
N Chow et al. Tracing the Evolutionary History and Global Expansion of Candida auris Using Population Genomic Analyses. mBio. DOI: : 10.1128/mBio.03364-19 (2020).
M Horton et al. Candida auris Forms High-Burden Biofilms in Skin Niche Conditions and on Porcine Skin. mSphere. DOI : 10.1128/mSphere.00910-19 (2020).
S Lockhard et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin Infect Dis. DOI: 10.1093/cid/ciw691 (2017)
Eix EF, CJ Johnson, KM Wartman, JF Kernien, JJ Meudt, D Shanmuganayagam, ALF Gibson, JE Nett. 2022. Ex vivo human and porcine skin effectively model C. auris colonization, differentiating robust and poor fungal colonizers. J Infect Dis. PMID: 35267041