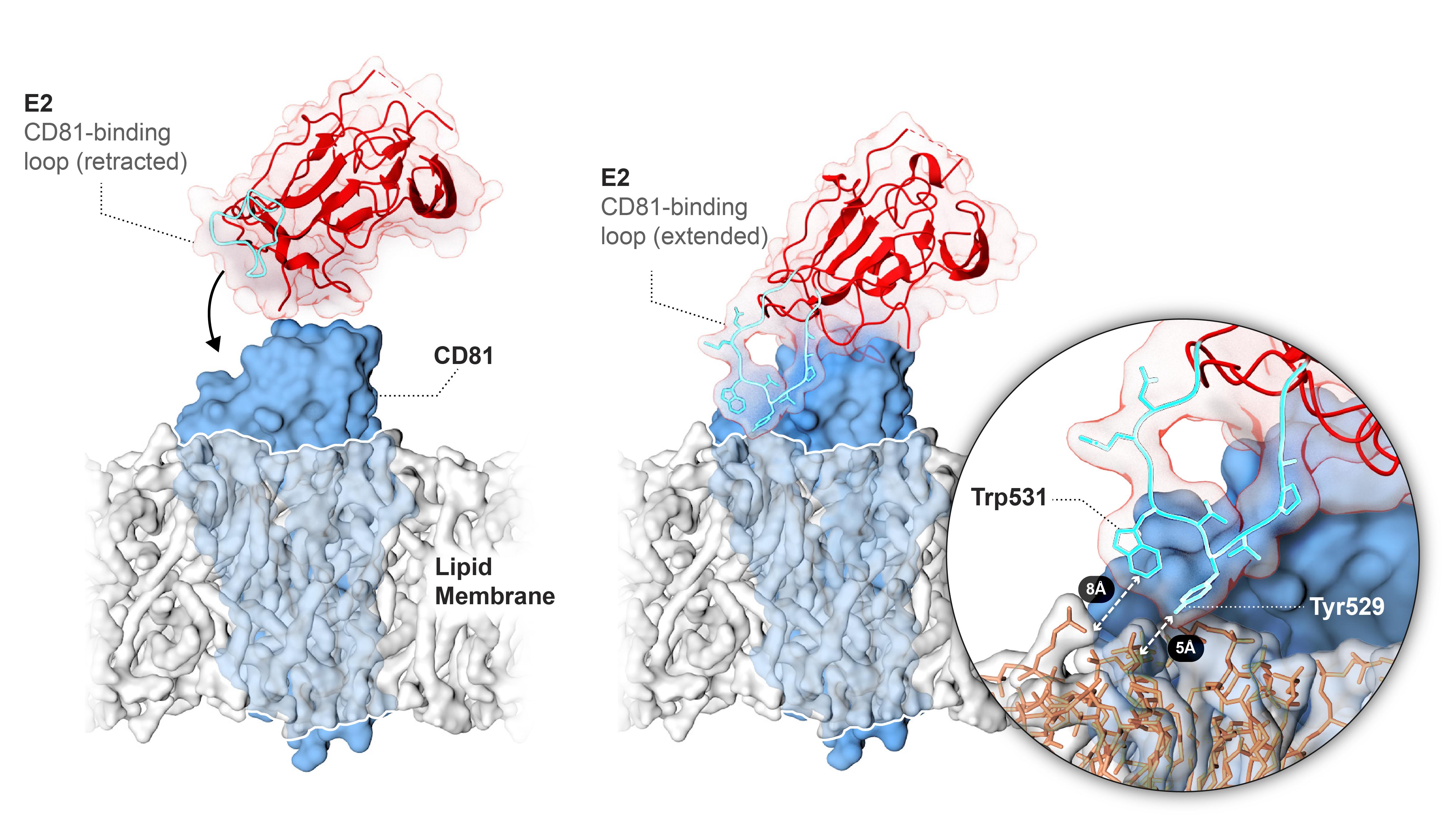
Melis M, Diaz G, Kleiner DE, Zamboni F, Kabat J, Lai J, Mogavero G, Tice A, Engle RE, Becker S, Brown CR, Hanson JC, Rodrigues-Canales J, Emmert-Buck M, Govindarajan S, Kew M, Farci P. Viral expression and molecular profiling in liver tissue versus microdissected hepatocytes in hepatitis B virus-associated hepatocellular carcinoma. J Transl Med. 2014 Aug 21;12:230.
Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, Farci P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013 Aug 15;133(4):816-24.
Farci P, Wollenberg K, Diaz G, Engle RE, Lai ME, Klenerman P, Purcell RH, Pybus OG, Alter HJ. Profibrogenic chemokines and viral evolution predict rapid progression of hepatitis C to cirrhosis. Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):14562-7.
Farci P, Diaz G, Chen Z, Govindarajan S, Tice A, Agulto L, Pittaluga S, Boon D, Yu C, Engle RE, Haas M, Simon R, Purcell RH, Zamboni F. B-cell gene signature with massive intrahepatic production of antibodies to hepatitis B core antigen in hepatitis B virus-associated acute liver failure. Proc Natl Acad Sci U S A. 2010 May 11;107(19):8766-71.
Farci P, Quinti I, Farci S, Alter HJ, Strazzera R, Palomba E, Coiana A, Cao D, Casadei AM, Ledda R, Iorio R, Vegnente A, Diaz G, Tovo PA. Evolution of hepatitis C viral quasispecies and hepatic injury in perinatally infected children followed prospectively. Proc Natl Acad Sci U S A. 2006 May 30;103(22):8475-80.
Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000 Apr 14;288(5464):339-44.
Visit PubMed for a complete publication listing.