Simmune Software Description
Simmune consists of three components:
- Simmune Modeler
- Simmune Cell Designer
- Simmune Simulator (for spatial simulations, a non-spatial version for fast model parameter scanning is forthcoming)
Here, we provide a brief overview of the Simmune package, which comes with extensive documentation and tutorials. Simmune is free for non-commercial use, and most of its source code will be made available shortly. Commercial licenses will be handled by the NIAID Technology Transfer and Intellectual Property Office.
Simmune Modeler
The Simmune modeler provides a visual interface for the definition of molecules, their structures (sub-molecular components or domains and binding sites), and the complexes the molecules form through reactions such as associations, dissociation, and transformations. Molecular reactions are mediated by interactions between individual binding sites. These interactions can be specified simply by drawing lines (using the mouse) connecting the binding sites. All reactions can be associated with state changes of the involved molecular complexes, providing straightforward, easily applied mechanisms to develop computational models of processes such as phosphorylations or other molecular modifications influencing the biochemical properties of cellular signaling components.
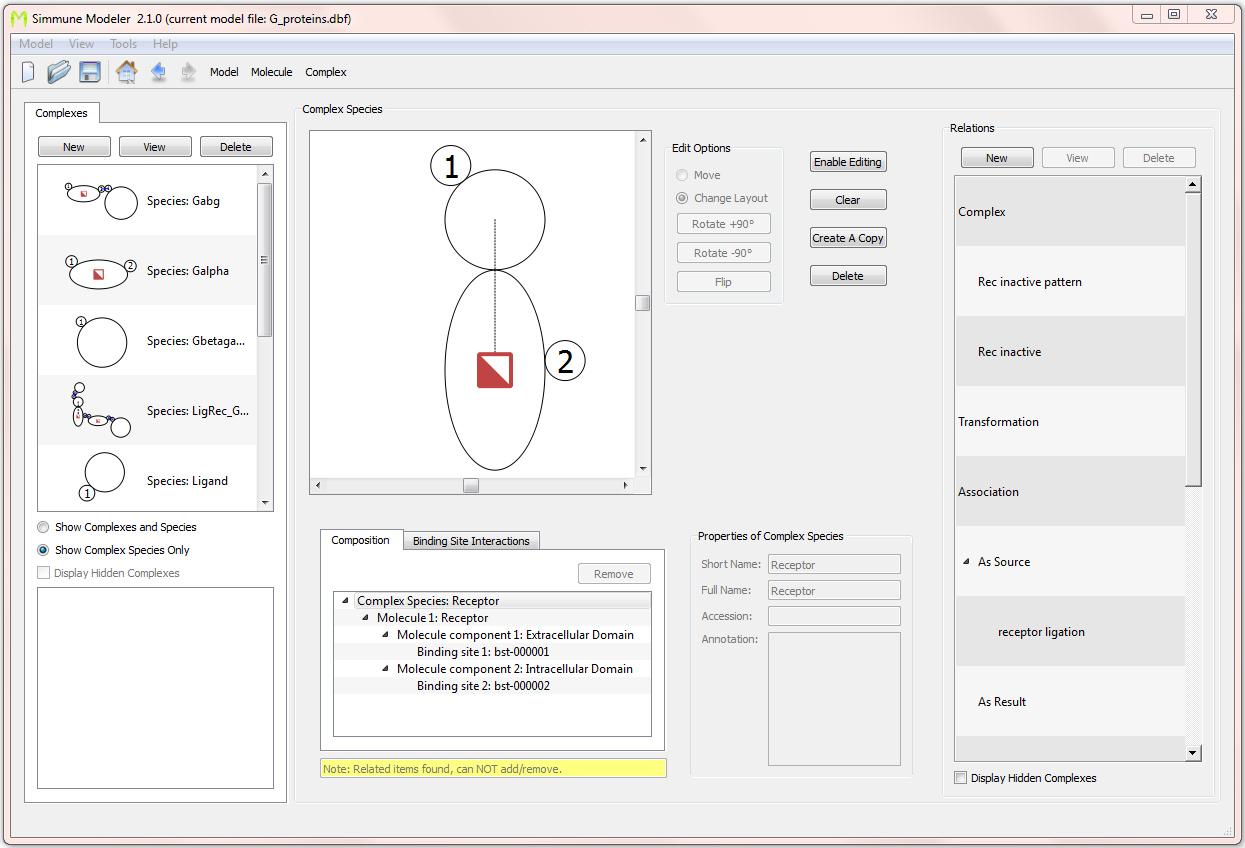
Screenshot of the interface for defining molecular complexes. Here, a single-molecular complex consisting of a trans-membrane receptor with one intra-cellular and one extra-cellular binding site is shown. The receptor has a state tag (depicted as a half-filled square) that is used for keeping track of its activation status. Defining such state tags and their functionality is done through an intuitive graphical process that does not require writing computer scripts.
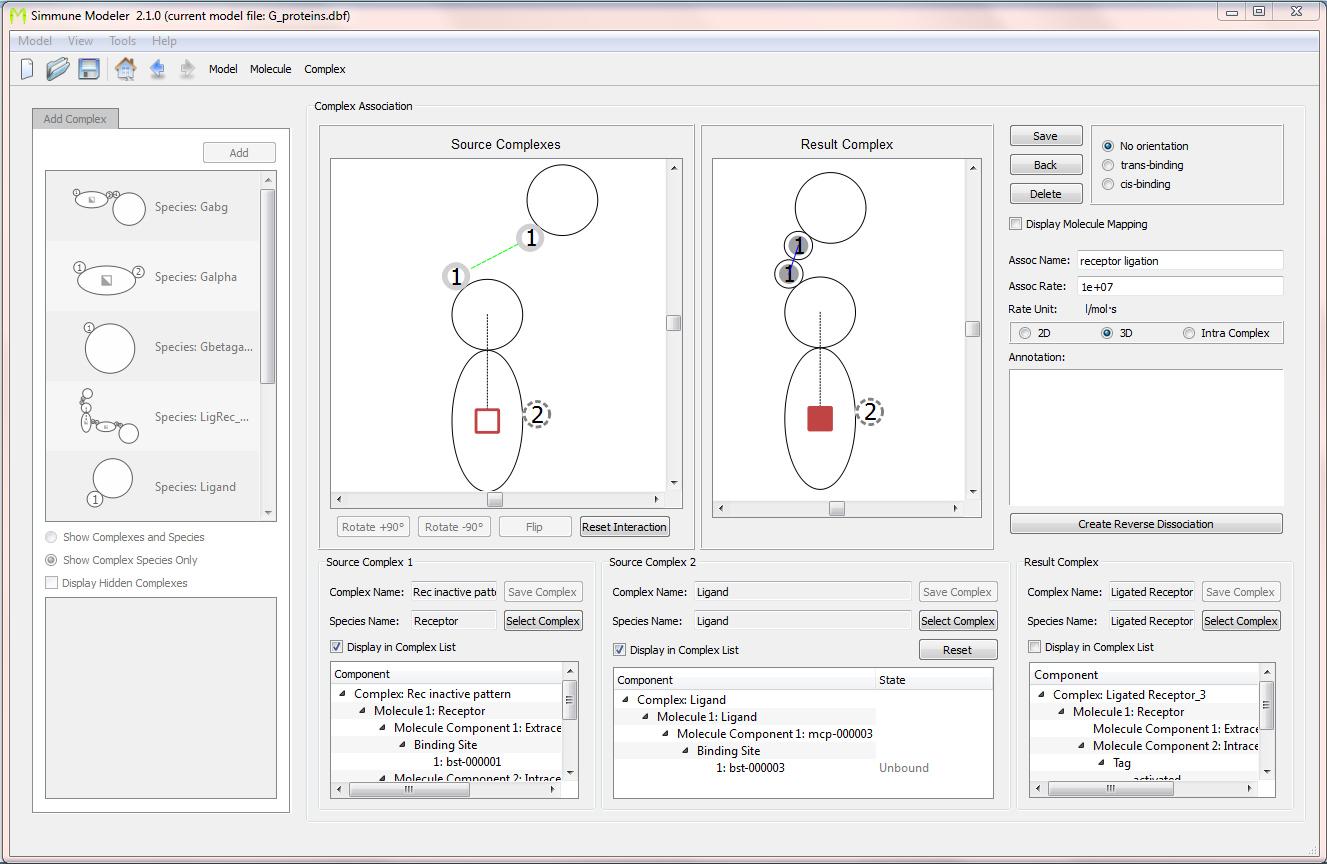
Screenshot of the interface for defining reactions (here: an association) between molecular complexes. The association between a receptor and its ligand is shown. The green dotted line connecting the extra-cellular binding site of the receptor and the (only) binding site of the ligand was drawn by mouse. The resulting complex was created simply by bringing the molecules into contact at the binding sites mediating the association. After right-clicking on the colored tag representing the activation state of the receptor, the initial state could be set to"'off" and the state of the ligated receptor to "on." Subsequently, the model was extended to specify binding between a G-protein alpha subunit and the receptor in its activated (but not its inactive) state.
Based on the definitions of such bi-molecular interactions, the software can automatically build the resulting network of all multimolecular complexes and their reactions. In the simple model of G-protein coupled receptor signaling for which two screenshots are shown here, a ligand-receptor association and an association between the activated receptor and a G-protein alpha subunit was specified. In a simulation of this system, the software would automatically take into account the three-molecule complex consisting of the ligand, the receptor, and the G-protein subunit if all three molecular species would be put into a simulated cell.
One unique feature of the Simmune software is its ability to build such reaction networks within the context of 3D cellular structures. Taking full advantage of the fact that this network generation proceeds automatically, the software can adapt the reactions even to cellular morphologies that change during simulations.
Simmune Cell Designer
The software provides a visual interface for the specification of cellular morphologies as well. These morphologies can be simply spherical or assume any (in principle) shape that may also include intracellular organelles. Additionally, the user can specify domains within these structures to create (initial) cellular biochemical contents with particular spatial characteristics.
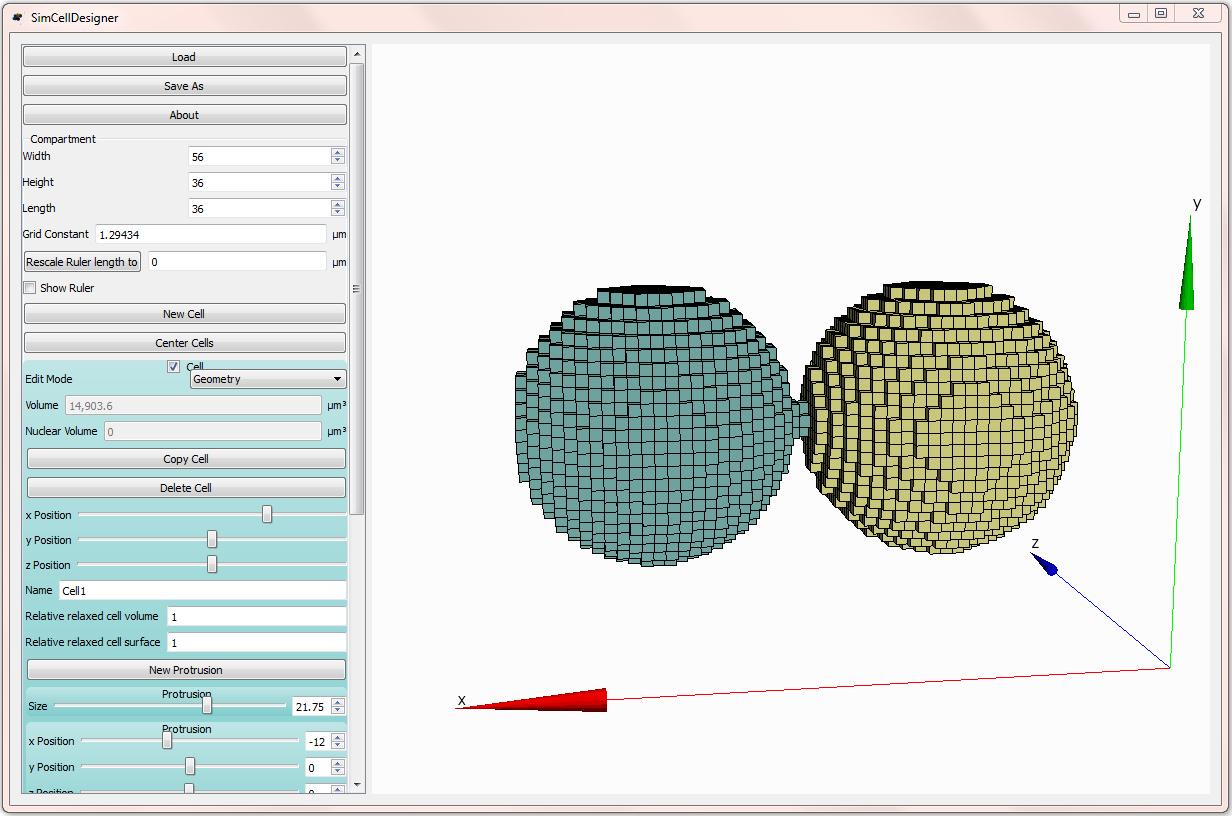
The graphical interface for defining cellular morphologies. Cells can be built in a modular way using combinations of “protrusions” that can be nested to specify intracellular organelles, embedded into cells of (in principle) any coherent shape.
For simulations with dynamic cellular morphologies, the user-defined shapes serve as initial conditions of the simulated cells.
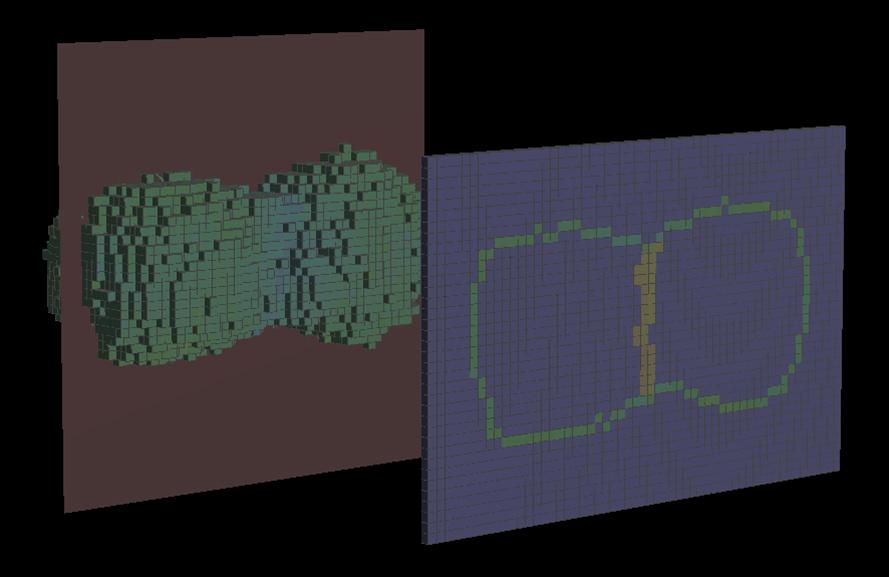
Evolution of an adhesion-driven cellular contact interface. In this simulation, the two cells were brought into contact initially at a small region of their membrane. Based on interactions between membrane-bound adhesion receptors, the cell contact then grew based on a Potts Model mechanism that performed stochastic updates of the cellular morphologies and had a lower probability for retracting volume elements carrying many intercellular adhesion complexes than for retracting other, less adhesive volume elements. Note that the software has to rebuild the local reaction networks of the interacting cadherin molecules automatically whenever a local change of the morphology takes place.
Simmune Simulator
After the molecular interactions and the (initial) cellular morphologies have been defined, the user can specify the initial biochemical contents of the cells to be simulated and then run simulations while inspecting the behavior of the cells with regard to, for example, the changing spatial distributions of molecular complexes or the total cellular contents of a particular molecule state (such as an activated receptor or a kinase) using a variety of tools.
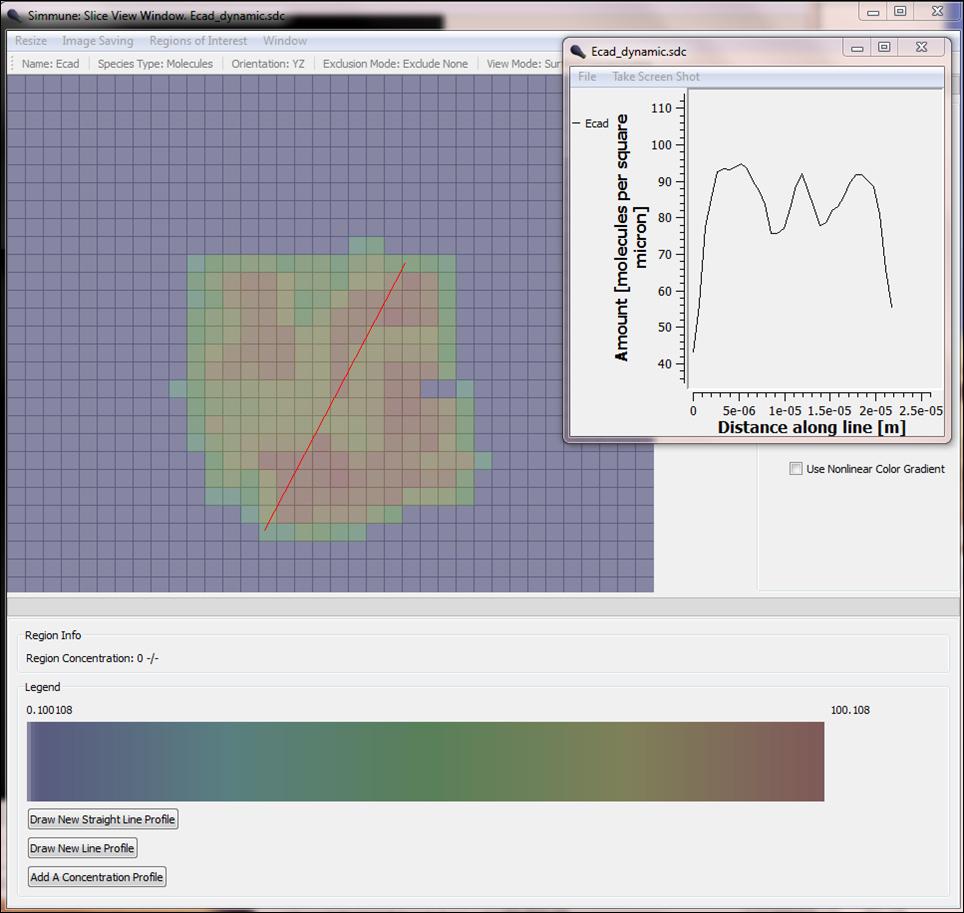
Concentration profile of E-cadherin adhesion receptors at the dynamic interface between two cells. The color-coded (red: high to blue: low) concentration of E-cadherin in a cut plane between two cells is shown. The inset shows the concentration along the red line.
Documentation of the simulator
Background
Simmune is the name of a suite of software tools that guides the user through the multiple hierarchical scales of cellular behavior, facilitating the generation of comprehensive models. It was originally created to simulate immunological phenomena—hence its name, Simmune—but it is applicable to a very broad class of cell biological models.
The development of Simmune started at the Institute for Theoretical Physics of the University of Hamburg, Germany. Now, its development continues as part of the biocomputation effort at the Laboratory of Immune System Biology.
The goal of our work is to provide the computational approaches appropriate for modeling cell biological systems based on data that describe cellular behavior on various scales, from interactions between molecular domains to the behavior of populations of cells.
In our view, such modeling software should not only offer cutting edge technical simulation capabilities but also, importantly, should make it possible for (experimental) biologists to define the computational equivalents of their biological models without having to deal with scripting languages or the mathematics involved in translating those models into formalized representations that can be (quantitatively) simulated.
The graphical user interface of the Simmune modeler allows the biologist to interact easily with the modeling software and largely eliminates the necessity to follow the classical strategy of first translating a biological model into a simplified version that then can be modeled by a theorist.
Other Software
2Pisa
Klauschen F, Ishii M, Qi H, Bajénoff M, Egen JG, Germain RN, Meier-Schellersheim M. Quantifying cellular interaction dynamics in 3D fluorescence microscopy data. Nat Protoc. 2009;4(9):1305-11.
- Software download file (ZIP)
Morphology Modeler (MoMo)
Klauschen F, Qi H, Egen JG, Germain RN, Meier-Schellersheim M Computational reconstruction of cell and tissue surfaces for modeling and data analysis. Nat Protoc. 2009;4(7):1006-12.
- Software download file (ZIP)
- Software ReadMe file (Word)

