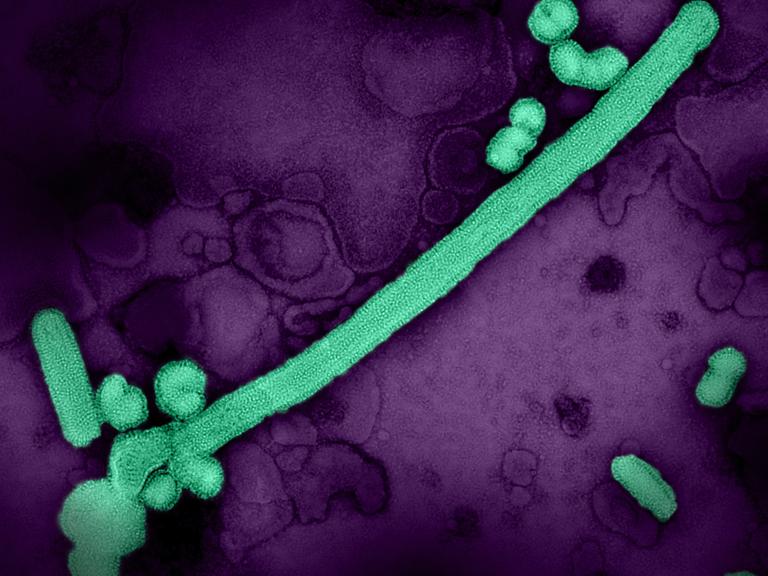
NIAID-funded researchers working in Peru have signaled concern about the deaths of birds and marine mammals from highly pathogenic avian influenza (HPAI) that has been spreading globally. Historically, South America’s coastal ecosystems and poultry industry have been spared from HPAI outbreaks.
“These viruses are rapidly accruing mutations, including mutations of concern, that warrant further examination and highlight an urgent need for active local surveillance to manage outbreaks and limit spillover into other species, including humans,” the researchers say.
Their study documenting the outbreak was published Sept. 7 in Nature Communications. It was led by Mariana Leguia of Pontificia Universidad Católica del Perú in Peru and EpiCenter for Emerging Infectious Disease Intelligence (Centers for Research in Emerging Infectious Diseases [CREID] Network). Other project collaborators included the National Forest and Wildlife Service of Peru; Wildlife Conservation Society; One Health Institute; and National Center for Biotechnology Information (NCBI) at the U.S. National Institutes of Health (NIH).
The study identifies flu type A/H5N1 lineage 2.3.4.4b, which in late 2021 spread from Europe to North America and then, in 2022, to South America. Along the way it has devastated wild birds and poultry farms. The scientists say their analyses supports a single introduction of 2.3.4.4b into Peru from North America during October 2022, presumably through the movements of migratory wild birds. The virus then infected local seabirds that share habitats with marine mammals.
Another recently published NIAID-funded study documents the evolution of the H5 virus over two decades from Europe to North America, prior to its spread to South America. The study—also in Nature Communications—involves St. Jude Children’s Research Hospital, part of NIAID’s Centers of Excellence for Influenza Research and Response (CEIRR) network.
In November 2022, scientists began documenting animal deaths along the Peruvian coast, including dolphins, sea lions, sanderlings, pelicans, and cormorants. The study—made possible by researchers quickly establishing new partnerships—identified HPAI A/H5N1 virus in the animals and analyzed how it had mutated since moving into South America.
They singled out one mutation to monitor: “… we are particularly concerned by the presence of PB2 D701N in 2 sea lion samples, and in a human case reported in Chile, as this mutation has been specifically linked to mammalian host adaptation and enhanced transmission.”
The project involved scientists obtaining 69 samples from 28 animals; they identified influenza A in 12 animals: a dolphin, four individual sea lions, one sample pooled from five sea lions, and six sea birds. The researchers then confirmed HPAI A/H5N1 in 11 of the 12 samples (one sample was too deteriorated to determine the specific type of influenza A). The researchers say there is concern of HPAI spreading to endangered species, such as the Andean condor, Humboldt penguin, and marine otter.
“An even larger concern,” they say, “is the possibility of spillover into human populations.” They say public awareness campaigns are needed to inform people about the presence of HPAI influenza and to avoid contact with animals that appear ill. The study notes that the outbreak in Peru occurred along the Pacific coast and during the summer, when many people go to the beach.
“It is not uncommon,” the study states, “for beachgoers (and their pets) to interact with sick and disoriented animals without any knowledge of the risks, or for free-roaming dogs in rural and semi-rural coastal areas to encounter sick or dead animals as they scavenge for food.”
In particular, they note, workers responsible for cleaning animal carcasses need additional training in the proper use of personal protective equipment and on waste management and disposal. “People in contact with sick and dead animals infected with HPAI A/H5N1 are at risk of infection,” the researchers say, “and human cases could be missed in the absence of active and obvious human-to-human transmission.”
These studies were funded in part by NIAID awards U01AI151814, 75N93021C00014, 75N93021C00016, HHSN272201400006C, and R01AI150745; and by the NIH's National Library of Medicine's Intramural Research Program.
References:
M Leguia, et al. Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Nature Communications DOI: 10.1038/s41467-023-41182-0 (2023).
A Kandeil, et al. Rapid evolution of A(H5N1) influenza viruses after intercontinental spread to North America. Nature Communications DOI: 10.1038/s41467-023-38415-7 (2023).