
Major Areas of Research
- Biology of antibody response to Plasmodium falciparum
- Characterization of human monoclonal antibodies to infectious pathogens
Program Description
Human monoclonal antibodies are emerging as powerful tools in combating infectious disease, both as direct prophylactics and as reagents to identify vulnerable sites on pathogens to guide vaccine design. At the Antibody Biology Unit (ABU), we aim to use cutting-edge technology to study B cells at the single cell level and to identify and characterize human monoclonal antibodies against a range of pathogens. We have two major aims:
- To study basic antibody biology. The sequences of monoclonal antibodies isolated from a vaccinated or naturally infected individual provide a high-resolution portrait of the antibody response to a given pathogen. Information revealed includes the predominant antibody isotype that is generated, the degree of somatic mutation and affinity maturation required for the development of a potent neutralizing response, and the preferential usage of specific VH genes to mount a response against a given antigen.
- To investigate the use of monoclonal antibodies for prevention of infection and as tools for vaccine design. Monoclonal antibodies that are isolated will be screened in in vitro and in vivo assays to determine their potency in preventing infection. Their affinity for their targets will be measured using biophysical assays. In collaboration with structural biologists, we will identify the specific epitopes targeted by the most potent antibodies and develop these sites as novel vaccine candidates. The most potent antibodies will also be considered as candidates to prevent infection in early-phase clinical trials.
The primary focus of the unit will be on malaria. Plasmodium falciparum causes approximately 400,000 deaths a year and remains a serious global health threat. Antibodies have been shown to be key mediators of protection against different stages of the P. falciparum life cycle, but the antibody response to malaria has only recently been studied at high resolution. The biology of the antibody response to P. falciparum is complex and fascinating. Recently, we identified broadly reactive antibodies from individuals living in malaria-endemic areas that contain a LAIR1 insert (an extra immunoglobulin-like domain) that is originally encoded in a different chromosome. This insert confers broad reactivity and is somatically mutated along with the rest of the antibody. This insertion event appears to be quite common in individuals living in different malaria-endemic regions (5-10% of individuals). In a separate study, we identified potent human monoclonal antibodies targeting a novel epitope on the P. falciparum circumsporozoite protein, the major sporozoite coat protein. This site is now being investigated as a new vaccine candidate.
Our platform is adaptable to any target. This unit will also study human monoclonal antibodies against other infectious agents, including Mycobacterium tuberculosis and SARS-CoV-2, as well as non-infectious targets.
Biography
Joshua Tan, Ph.D., is a Stadtman Tenure-track Investigator and an NIH Distinguished Scholar in the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases. He received his Ph.D. from the University of Oxford, England. Prior to joining the NIH, he was awarded the Pfizer Research Prize for his malaria work and the Sir Henry Wellcome Postdoctoral Fellowship to investigate human monoclonal antibodies that target the malaria-causing parasite P. falciparum.
Research Group
Andrew Cooper, Ph.D., Postdoctoral Fellow
Cherrelle Dacon, Ph.D., Postdoctoral Fellow
Divya Mohan, Biologist
Lauren Purser, Lab Manager
Lawrence Wang, Visiting PhD Student
Courtney Tucker, Ph.D. Student
Selected Publications
Cho H, Gonzales-Wartz KK, Huang D, Yuan M, Peterson M, Liang J, Beutler N, Torres JL, Cong Y, Postnikova E, Bangaru S, Talana CA, Shi W, Yang ES, Zhang Y, Leung K, Wang L, Peng L, Skinner J, Li S, Wu NC, Liu H, Dacon C, Moyer T, Cohen M, Zhao M, Lee FE, Weinberg RS, Douagi I, Gross R, Schmaljohn C, Pegu A, Mascola JR, Holbrook M, Nemazee D, Rogers TF, Ward AB, Wilson IA, Crompton PD and Tan J. 2021 Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci Transl Med 13, eabj5413.
Tan J, Cho H, Pholcharee T, Pereira LS, Doumbo S, Doumtabe D, Flynn BJ, Schön A, Kanatani S, Aylor SO, Oyen D, Vistein R, Wang L, Dillon M, Skinner J, Peterson M, Li S, Idris AH, Molina-Cruz A, Zhao M, Olano LR, Lee PJ, Roth A, Sinnis P, Barillas-Mury C, Kayentao K, Ongoiba A, Francica JR, Traore B, Wilson IA, Seder RA and Crompton PD. 2021. Functional human IgA targets a conserved site on malaria sporozoites. Sci Transl Med 13, abg2344.
Tan J, Piccoli L and Lanzavecchia A. 2019. The antibody response to Plasmodium falciparum: cues for vaccine design and the discovery of receptor-based antibodies. Annu Rev Immunol 37, 225-246.
Tan J, Sack BK, Oyen D, Zenklusen I, Piccoli L, Barbieri S, Foglierini M, Fregni CS, Marcandalli J, Jongo S, Abdulla S, Perez L, Corradin G, Varani L, Sallusto F, Sim BKL, Hoffman SL, Kappe SHI, Daubenberger C, Wilson IA and Lanzavecchia A. 2018. A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein. Nat Med 24, 401-407.
Pieper K, Tan J, Piccoli L, Foglierini M, Barbieri S, Chen Y, Fregni CS, Wolf T, Jarrossay D, Anderle M, Abdi A, Ndungu FM, Doumbo OK, Traore B, Tran TM, Jongo S, Zenklusen I, Crompton PD, Daubenberger C, Bull PC, Sallusto F and Lanzavecchia A. 2017. Public antibodies to malaria antigens generated by two LAIR1 insertion modalities. Nature 548, 597-601.
Tan J, Pieper K, Piccoli L, Abdi A, Foglierini M, Geiger R, Tully CM, Jarrossay D, Ndungu FM, Wambua J, Bejon P, Fregni CS, Fernandez-Rodriguez B, Barbieri S, Bianchi S, Marsh K, Thathy V, Corti D, Sallusto F, Bull P and Lanzavecchia A. 2016. A LAIR1 insertion generates broadly reactive antibodies against malaria variant antigens. Nature 529, 105-109.
Highlight
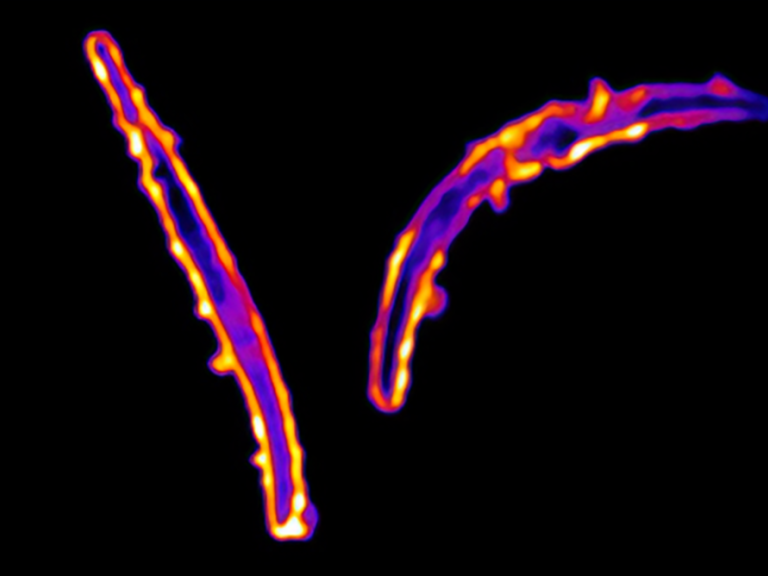
NIH Researchers Discover Novel Class of Anti-Malaria Antibodies
January 3, 2025
New antibodies that bind to a previously untargeted portion of the malaria parasite could lead to new monoclonal antibody treatments and vaccines for malaria.
Human monoclonal antibodies are emerging as powerful tools to combat infectious disease. At the Antibody Biology Unit (ABU), we aim to use cutting-edge technology to study B cells at the single-cell level and to identify and characterize human monoclonal antibodies against a range of pathogens. The Unit currently focuses on diseases with substantial burden globally or in the United States.
ABU has two main objectives:
- Investigate the use of monoclonal antibodies for the prevention of infection and as tools for immunogen design: An array of tools, including the Berkeley Lights Beacon and Carterra LSA, will be used for high-throughput antibody identification and characterization. Monoclonal antibodies that are isolated will be screened in in vitro and in vivo assays to determine their potency in preventing infection. Their affinity for their targets will be measured using biophysical assays. In collaboration with structural biologists, we will identify the specific epitopes targeted by the most potent antibodies and develop these sites as novel immunogens. The most potent antibodies will also be considered as candidates to prevent infection in early-phase clinical trials.
- Study basic antibody biology: The sequences of monoclonal antibodies isolated from an immunized or naturally infected individual provide a high-resolution portrait of the antibody response to a given pathogen. Information revealed includes the predominant antibody isotype that is generated, the degree of somatic mutation and affinity maturation required for the development of a potent neutralizing response, and the preferential usage of specific VH genes to mount a response against a given antigen.
Dacon C, Moskovitz R, Swearingen K, Da Silva Pereira L, Flores-Garcia Y, Aleshnick M, Kanatani S, Flynn B, Molina-Cruz A, Wollenberg K, Traver M, Kirtley P, Purser L, Dillon M, Bonilla B, Franco A, Petros S, Kritzberg J, Tucker C, Paez GG, Gupta P, Shears MJ, Pazzi J, Edgar JM, Teng AA, Belmonte A, Oda K, Doumbo S, Krymskaya L, Skinner J, Li S, Ghosal S, Kayentao K, Ongoiba A, Vaughan A, Campo JJ, Traore B, Barillas-Mury C, Wijayalath W, Idris A, Crompton PD, Sinnis P, Wilder BK, Zavala F, Seder RA, Wilson IA, Tan J. Protective antibodies target cryptic epitope unmasked by cleavage of malaria sporozoite protein. Science. 2025 Jan 3;387(6729):eadr0510.
Wang LT, Cooper AJR, Farrell B, Miura K, Diouf A, Müller-Sienerth N, Crosnier C, Purser L, Kirtley PJ, Maciuszek M, Barrett JR, McHugh K, Ogwang R, Tucker C, Li S, Doumbo S, Doumtabe D, Pyo CW, Skinner J, Nielsen CM, Silk SE, Kayentao K, Ongoiba A, Zhao M, Nguyen DC, Lee FE, Minassian AM, Geraghty DE, Traore B, Seder RA, Wilder BK, Crompton PD, Wright GJ, Long CA, Draper SJ, Higgins MK, Tan J. Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5. Cell. 2024 Sep 5;187(18):4981-4995.e14.
Dacon C, Peng L, Lin TH, Tucker C, Lee CD, Cong Y, Wang L, Purser L, Cooper AJR, Williams JK, Pyo CW, Yuan M, Kosik I, Hu Z, Zhao M, Mohan D, Peterson M, Skinner J, Dixit S, Kollins E, Huzella L, Perry D, Byrum R, Lembirik S, Murphy M, Zhang Y, Yang ES, Chen M, Leung K, Weinberg RS, Pegu A, Geraghty DE, Davidson E, Doranz BJ, Douagi I, Moir S, Yewdell JW, Schmaljohn C, Crompton PD, Mascola JR, Holbrook MR, Nemazee D, Wilson IA, Tan J. Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses. Cell Host Microbe. 2023 Jun 14;31(6):1071-1072.
Dacon C, Tucker C, Peng L, Lee CD, Lin TH, Yuan M, Cong Y, Wang L, Purser L, Williams JK, Pyo CW, Kosik I, Hu Z, Zhao M, Mohan D, Cooper AJR, Peterson M, Skinner J, Dixit S, Kollins E, Huzella L, Perry D, Byrum R, Lembirik S, Drawbaugh D, Eaton B, Zhang Y, Yang ES, Chen M, Leung K, Weinberg RS, Pegu A, Geraghty DE, Davidson E, Douagi I, Moir S, Yewdell JW, Schmaljohn C, Crompton PD, Holbrook MR, Nemazee D, Mascola JR, Wilson IA, Tan J. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science. 2022 Jul 12:eabq3773.
Cho H, Gonzales-Wartz KK, Huang D, Yuan M, Peterson M, Liang J, Beutler N, Torres JL, Cong Y, Postnikova E, Bangaru S, Talana CA, Shi W, Yang ES, Zhang Y, Leung K, Wang L, Peng L, Skinner J, Li S, Wu NC, Liu H, Dacon C, Moyer T, Cohen M, Zhao M, Lee FE, Weinberg RS, Douagi I, Gross R, Schmaljohn C, Pegu A, Mascola JR, Holbrook M, Nemazee D, Rogers TF, Ward AB, Wilson IA, Crompton PD, Tan J. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci Transl Med. 2021 Oct 20;13(616):eabj5413.
Tan J, Pieper K, Piccoli L, Abdi A, Perez MF, Geiger R, Tully CM, Jarrossay D, Maina Ndungu F, Wambua J, Bejon P, Fregni CS, Fernandez-Rodriguez B, Barbieri S, Bianchi S, Marsh K, Thathy V, Corti D, Sallusto F, Bull P, Lanzavecchia A. A LAIR1 insertion generates broadly reactive antibodies against malaria variant antigens. Nature. 2016 Jan 7;529(7584):105-109.
- Development of monoclonal antibody therapeutics against pathogens
- Characterization of human antibody response to infectious pathogens
- Biology of antibody response to influenza, other viruses, Plasmodium falciparum, and Mycobacterium tuberculosis
Systems Immunology of COVID-19
*This project is not currently being actively pursued.
Participating Hospitals
Brescia Civil Hospital
Brescia, Italy
Principal Investigators: Luisa Imberti and Eugenia Quiros
Children's Hospital of Philadelphia
Philadelphia, Pennsylvania, USA
Principal Investigator: Sarah Henrickson
Cytokine Profile
Project Title: IFN signature response and cytokine profiling of samples from hospitalized patients with respiratory syndrome coronavirus 2 (SARS-CoV-2) disease (COVID-19)
NIAID Principal Investigator: Raphaela Goldbach-Mansky M.D., M.H.S.
Chief, Translational Autoinflammatory Diseases Section, LCIM
SARS-CoV-2 Antibody Response
Project Title: Analysis of SARS-CoV-2-specific antibody responses
NIAID Principal Investigator: Jeffrey Cohen M.D.
Chief, Laboratory of Infectious Diseases
Chief, Medical Virology Section, LID
TCR and BCR Repertoire
Project Title: Composition of T- and B-Cell Repertoire and Mapping of Virus-Specific T-Cell Receptor Sequences
NIAID Principal Investigator: Luigi Notarangelo, M.D.
Chief, Laboratory of Clinical Immunology and Microbiology
Chief, Immune Deficiency Genetics Section, LCIM
Host Genetics
Project Title: Genetic Determinants of Susceptibility to Severe COVID-19 Infection
NIAID Principal Investigator: Helen Su, M.D., Ph.D
Chief, Human Immunological Diseases Section, LCIM
Immune Response to COVID-19
Comprehensive analyses of innate and adaptive immune responses during acute COVID-19 infection and convalescence
The COVID-19 pandemic poses an unprecedented public health crisis. At present, our narrow understanding of the immune system’s response to the infection limits our capacity to prevent and treat severe disease. As part of the efforts outlined in the NIAID Strategic Plan for COVID-19 Research, NIAID researchers are spearheading a large, international collaboration to unveil the innate and adaptive immune responses during acute COVID-19 infection and convalescence. Each researcher will contribute their unique expertise to collectively elucidate the innate and adaptive immune response to COVID-19 infection. This synergistic coalition of researchers will work closely and share data to maximize the impact of patient samples. The overall goal is to identify immunological and virological correlates and predictors of clinical outcomes.
The research projects will examine the following:
- Genetic markers of susceptibility to severe COVID-19 infection
- Composition of T- and B-cell repertoire and mapping of virus-specific T-cell receptor (TCR) sequences
- Cytokine and chemokine profiling, including interferon (IFN) signature and soluble markers of inflammation
- Antibody responses to COVID-19 infection
- Anti-cytokine autoantibodies
- Levels of plasma gelsolin
- Humoral immunological signature of the human virome
- Anti-commensal antibody repertoire
- Systems biology approach to understand changes in the immune system
- Intrapatient SARS-CoV-2 genetic variation
- Role of neutrophil extracellular traps (NETs)
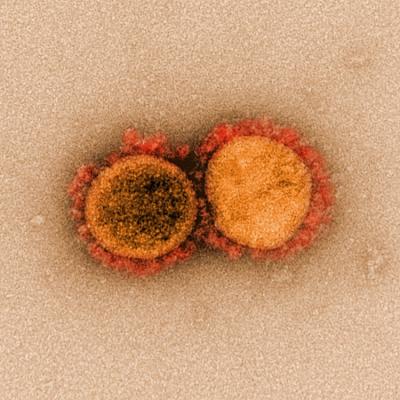
Transmission electron micrograph of SARS-CoV-2 virus particles, isolated from a patient.
Researchers in the Human Immunological Diseases Section believe COVID-19 causes mild or no illness in some individuals and not others because of our genetic makeup. The lab will sequence and analyze the genomes of previously healthy patients who experienced severe or fatal COVID-19 infection.
The Immune Deficiency Genetics Section aims to analyze the dynamic changes that occur within an individual’s T-cell and B-cell repertoires during the transition from acute COVID-19 infection into convalescence.
The Translational Autoinflammatory Diseases Section will harness the power of Nanostring and RNA-Seq to measure and characterize the pro-inflammatory cytokine signature of COVID-19.
The Medical Virology Section is interested in performing serologic studies to better understand the epidemiology of COVID-19 in specific cohorts.
Researchers in the Immunopathogenesis Section believe that anti-cytokine antibodies that can neutralize antiviral cytokines may contribute to COVID-19 disease progression, as they do in other immune-mediated diseases.
The NIAID Microbiome Program and the Metaorganism Immunity Section propose that immune responses to the gut microbiota may predict which patients get very sick with COVID-19 while others remain asymptomatic.
The Multiscale Systems Biology Section will integrate computational approaches and several cutting-edge high-throughput methods to assess changes in the epigenome, transcriptome, and proteome throughout the course of COVID-19 disease.
The Virus Persistence and Dynamics Section will explore the intrapatient diversity of SARS-CoV-2 sequences using high-throughput, long-read, single-molecule sequencing.
The Systemic Autoimmunity Branch at NIAMS will study in detail the role of neutrophil dysregulation and NETs in the induction of enhanced proinflammatory responses, tissue damage, vasculopathy, and prothrombotic manifestations.
We seek hospitals to collaborate in our research project by submitting samples. For sample requirements, see the specific project pages.
These are original research publications using samples obtained from the NIAID COVID Consortium. Each paper addresses one or more of the scientific questions outlined in the project pages, with the goal of elucidating the immune response to COVID-19.
Evaluation of Patients With Immune Function Abnormalities
Contact Information
Patricia L Littel, R.N., (301) 402-5964
plittel@cc.nih.gov
Harry L Malech, M.D., (301) 480-6916
hmalech@nih.gov
Use of G-CSF to Obtain Blood Cell Precursors
Contact Information
Contact: Joanna L Peterson, (240) 760-6506
joanna.peterson@nih.gov
Contact: Harry L Malech, M.D., (301) 480-6916
hmalech@nih.gov

