Davis-Gardner ME et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA Bivalent Booster. NEJM, 2022 December
Wang Q et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell, 2022 December


Davis-Gardner ME et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA Bivalent Booster. NEJM, 2022 December
Wang Q et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell, 2022 December
The ability of NIAID to prepare for and mount a rapid and effective research response to an emerging pathogen relies on knowledgeable staff, up-to-date facilities and flexible infrastructure, cutting-edge technology, and a robust, centralized coordination hub.
NIAID will leverage its expertise in infectious diseases, genomics, proteomics, bioinformatics, and access to clinical samples to develop rapid-response diagnostics for biological threats and emerging infectious diseases. In close collaboration with other NIH institutes and USG agencies, NIAID will support the development new and improved point-of-care and home-based tests as well as ultrahigh-throughput central reference laboratory testing that can accurately detect signatures of infectious pathogens that are of high public-health consequence.
Developing products that can protect against pathogens of concern is an integrated process that requires basic and applied research. Fundamental knowledge of pathogen biology; structural properties; mechanisms of transmission, including identification of viral vectors; viral lifecycle; viral entry mechanisms and host receptors; tissue tropism; and host immune responses is critical to efforts informing the development of MCMs against new, emerging, or remerging pathogens.
The NIAID Pandemic Preparedness Plan builds on a foundation of pathogen-specific research that include advancing research on priority pathogens known to or having the potential for emerging as public health threats. Continuing to build a robust basic research portfolio and advancing translational science on these pathogens is essential for biomedical preparedness. In addition to known threats, effective preparedness must also account for unexpected emerging disease threats, commonly referred to as Pathogen(s) X.
The African Centers of Excellence in Bioinformatics and Data-Intensive Science (ACE) are a consortium of research and training centers facilitated by the National Institute of Allergy and Infectious Diseases (NIAID) in collaboration with African research institutions, private sector companies, and the Foundation for the NIH. ACE delivers high performance computing infrastructure and training through a public-private partnership based on in-kind contributions. ACE also provide researchers with mentoring, training, technical and scientific support, computer resources, and the bioinformatics tools necessary for advanced data analysis. Finally, ACE provides a platform for sharing skills, knowledge, and best practices between members of the global biomedical research community.
Read more about this network: African Centers of Excellence in Bioinformatics and Data-Intensive Science

ACE Senior Engineer Rodgers Kimera in virtual reality (VR) headset.
The SARS-CoV-2 Interagency Group (SIG), established by the U.S. Department of Health and Human Services (HHS), works to rapidly characterize emerging variants and actively monitors their potential impact on SARS-CoV-2 vaccines, therapeutics, and diagnostics. The SIG is responsible for variant classifications in the United States and meets regularly to evaluate the risk posed by SARS-CoV-2 variants circulating in the United States and globally to make recommendations about the variants. NIAID is a member of the SIG and supports research and surveillance activities to help inform decisions made by the SIG.
To support the generation of data for the SIG, NIAID has created the SARS-CoV-2 Assessment of Viral Evolution (SAVE) risk-assessment pipeline for SARS-CoV-2 variant viruses. SAVE provides a comprehensive real-time risk assessment of emerging mutations in SARS-CoV-2 that could impact transmissibility, virulence, and susceptibility to infection- or vaccine-induced immunity. The goals of the SAVE program are to understand emerging changes in the virus that could impact transmissibility, virulence, susceptibility to convalescent and vaccine-induced immunity and diagnostic testing. SAVE is one of the data streams that provides important information for variant characterization and complements other USG and NIH efforts like the ACTIV Tracking Resistance and Coronavirus Evolution (TRACE) initiative.
The NIAID SAVE program is composed of an international team of scientists with expertise in virology, immunology, vaccinology, structural biology, bioinformatics, viral genetics, and evolution. Researchers from NIAID Intramural, the NIAID Vaccine Research Center, other HHS and Department of Defense laboratories, and the extramural academic community work collaboratively within and across multiple sub-groups to accelerate the pace of variant research and discovery through rapid and open sharing. Researchers are developing high quality data sets that are then vetted by SIG working groups comprised of various experts. These data help inform public health recommendations.
More information on the SAVE program can be found in the publication, "Defining the risk of SARS-CoV-2 variants on immune protection" (Nature, 2022).
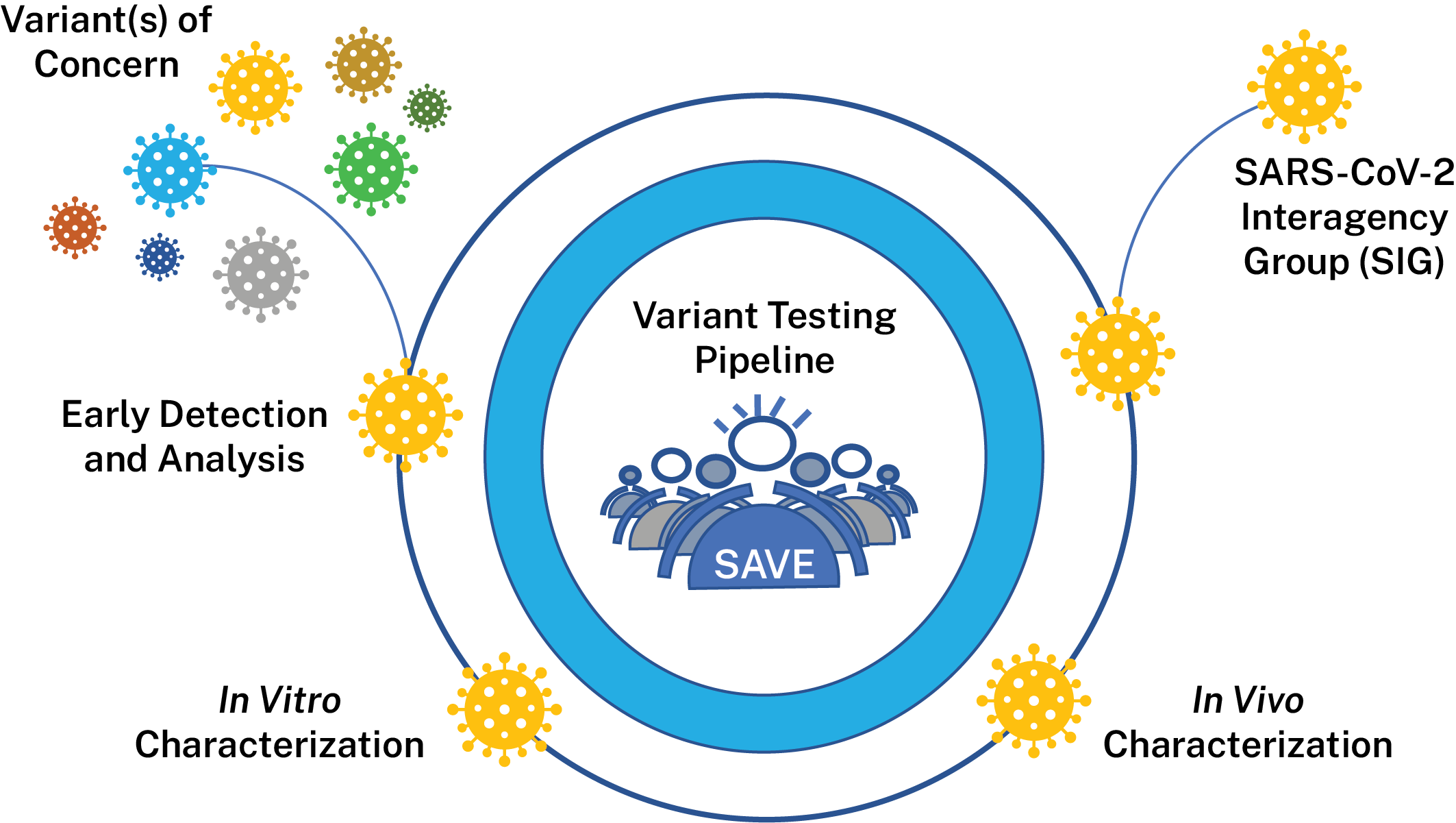
NIAID’s activities are divided into the following data-generating components:
Lead: Elodie Ghedin, Ph.D., NIAID and Derek Smith, Ph.D., Cambridge University
This team is evaluating surveillance/epidemiological data, information from observational studies on the efficacy of COVID-19 vaccines, variant information from documented vaccine breakthrough cases and accompanying sequence data from these efforts to identify variants that should be evaluated further in vitro and in vivo. They will also perform antibody escape mapping in the lab using monoclonal antibodies and polyclonal sera in pseudoviral assays to identify proactively substitutions of concern.
Lead: Florian Krammer, Ph.D., Icahn School of Medicine at Mount Sinai and Mehul Suthar, Ph. D., Emory University
This team characterizes variants identified by the early detection group comprehensively in vitro to determine how they may differ from the original SARS-CoV-2 virus. Variant viruses, pseudoviruses and various convalescent and vaccinee sera samples are used to determine if variants are able to evade infection- or vaccine-induced immunity. Groups are also performing antigenic landscape analyses to determine the relatedness of each emerging variant to the original strain to which vaccines were made. Researchers also analyze characteristics such as in vitro replication kinetics to determine how variant viruses may have different characteristics that affect their ability to replicate or transmit between humans. Groups are also exploring if any variants can evade cellular immune responses generated by memory B-cells and T-cells. The data generated by this groups helps determine which variants are of most concern for our existing countermeasures.
Lead: Michael Diamond, M.D., Ph.D., and Jacco Boon, Ph.D., Washington University in St. Louis
This team is using animal models to estimate virulence changes in circulating SARS-CoV-2 viruses and evaluate vaccine efficacy against variant viruses. Researchers use various animal models to examine if any variants cause different levels of disease in models. Researchers also vaccinate or infect animals to determine if animals remain protected from infection with variant viruses over different periods of time. Finally, researchers are working to study ability of countermeasures such as vaccines to prevent transmission in animal models.
SAVE is generating and characterizing reagents to be used across the USG and the global scientific community, including sera panels, viruses and protein. SAVE-generated and other available resources can be found at BEI Resource Repository.
SAVE has been involved in the research published in a variety of scientific journals focusing recently on the Omicron and Delta variants.
Federal agencies and laboratories, academic institutions, and corporations collaborate to make SAVE possible.
Office: NIH Office of Patient Recruitment
Phone: (800) 411-1222
TTY: (866) 411-1010
Email: ccopr@nih.gov