Structural Virology and Vaccinology Section
Tongqing Zhou, Ph.D.
Contact: tzhou@mail.nih.gov

Major Areas of Research
- Structure-based vaccine design
- Neutralizing antibodies
- Atomic-level understanding of the HIV-1 envelope and its evasion from the immune system
Program Description
The Structural Virology and Vaccinology Section (SVVS) seeks to apply structural biology to the development of effective vaccines — especially a vaccine against HIV-1.
One area in which we and others have already made an impact is in understanding how HIV-1 is able to evade the humoral immune system. Determination of the structure of the HIV-1 gp120 envelope glycoprotein (Kwong 1998 Nature 393, 648-659), provided a physical map of the primary target of neutralizing antibodies against HIV-1 and showed how gp120 conformational diversity can prevent antibody-mediated neutralization (Kwong 2002 Nature 420, 678-682) andhow N-linked carbohydrates can form an "evolving glycan shield" (Wei 2003 Nature 422, 307-312). These and other studies, including the determination of crystal structures of the entire spike ectodomain (Pancera 2014 Nature 514, 455-461) and the determination of crystal structures of fully glycosylated envelope trimers (Stewart-Jones 2016 Cell 165, 813-826) — have led to an understanding of the molecular trickery that protects HIV-1 from the humoral immune response.
A second area in which we and others have made an impact is in understanding how human antibodies can neutralize diverse HIV-1 isolates. The structure of the b12 antibody revealed how the binding site for the CD4 receptor formed a site of vulnerability to neutralizing antibody (Zhou 2007 Nature 329, 811-817).
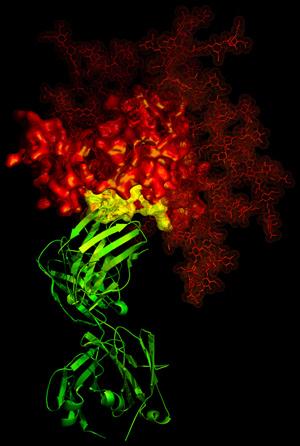
3-D X-ray crystallographic image showing the broadly neutralizing antibody b12 (green ribbon) in contact with a critical target (yellow) for vaccine developers on HIV-1 gp120 (red). Read more about this discovery.
The VRC01 antibody, which we participated in identifying in 2010 (Wu 2010 Science 329, 856-861), uses mimicry of the CD4 receptor to neutralize over 90 percent of HIV-1 isolates, though the inability of germline versions of VRC01 to bind to HIV-1 Env and its extraordinary level of somatic hypermutation suggest roadblocks to eliciting similar antibodies (Zhou 2010 Science 329, 811-817). Importantly, the Antibody Mediated Prevention (AMP) study (HVTN 704/HPTN 085) showed that an IC80 of less than 1 µg/ml was required for protection by antibody VRC01. We (and the field) are following up on the implications of this study, but high breadth and potency are clearly required. We have succeeded in improving the breadth and potency of HIV-1-neutralzing antibodies so that the best, such as VRC01.23 (Kwon 2021 Mabs 13:1946918), can neutralize over 90% of our 208-strain panel with an IC80 of less than 1 µg/ml.
But can one use structural biology in vaccine design? Currently, we are following three lines of investigation.
One line involves understanding the B-cell pathways that produce broadly neutralizing antibodies and seeking to replicate their development. We’ve observed select broadly neutralizing antibodies to develop similarly in multiple donors (e.g., Wu 2011 Science 333, 1593-1602; Zhou 2013 Immunity 39, 245-258; Bonsignori 2011 J. Virol. 85, 9998-10009; Doria-Rose 2014 Nature 509, 55-62) suggesting that — for select antibodies — a common set of immunogens might spur the induction and maturation of similar antibodies in the general population.
A second line involves the precise delineation of functional constraints to identify potential footholds of conservation and exposure. One functional constraint involves receptor binding — with the site on HIV-1 Env involved in binding the CD4 receptor providing a “supersite of vulnerability.” Analysis of recognition of the CD4 supersite in 14 donors suggests that steric access to the CD4 supersite is a primary physical constraint limiting antibody recognition (Zhou 2015 Cell 161, 1280-1292). Another functional constraint involves the viral fusionmachinery, including the fusion peptide, which grabs hold of the target cell membrane as an essential step in virus entry. We've found the N-terminal eight residues of the fusion peptide to be a region targeted by neutralizing antibodies (Kong 2016 Science 352, 828-338).
A third line involves the structure-based engineering of the trimeric spike ectodomain into immunogens with the ability to stimulate the induction of broadly neutralizing antibodies. The success of this line of investigation depends in part on the design of spike mimics, which are specific for broadly neutralizing antibodies and not recognized by the non- or poorly-neutralizing antibodies that typically dominate the humoral immune response; and we have used structure-based design to produce prefusion-stabilized HIV-1 Env spikes of appropriate antigenicity from a variety of Env strains (Kwon 2015 NSMB 22, 522-531; Joyce 2017 Cell Rep. 21, 2992-3002; Rutten 2018 Cell Rep. 23, 584-595).
Excitingly, by combining both the second and third lines of investigation, we have succeeded in inducing cross-clade neutralizing antibodies in mice, guinea pigs, and rhesus macaques (Xu 2018 Nat Med. 24, 857-867). Specifically, we’ve used prefusion-stabilized Env spikes to boost fusion peptide-directed responses induced by vaccinating with fusion peptide coupled to the highly immunogenic carrier, keyhole limpet hemocyanin (KLH), a standard workhorse of the monoclonal antibody industry. We have now manufactured a fusion peptide priming reagent, using the heavy chain of tetanus toxoid (Ou 2019 Sci Rep 10:3032), and are initiating clinical trials to assess the ability of immunogens to develop broadly neutralizing antibodies in humans.
Thus, by combining multiple approaches, including those used in biotechnology, we have been able to elicit broadly neutralizing antibodies against HIV-1, especially against the fusion peptide site of vulnerability. Complete functional screening indicates resistance to these antibodies to be focused almost exclusively to the N-terminus of the fusion peptide (Dingens 2018 PLoS Pathogens 14:e1007159), a focus obtained through somatic hypermutation during the priming phase (Kong 2019 Cell 178, 567-584).
While structure-based vaccine development with HIV-1 is proceeding, we have also been working to test our structure-based approach with other viral pathogens. We have engineered a promising vaccine antigen against respiratory syncytial virus (RSV), the leading cause of hospitalization for children under five years of age. Our “conformational fixation” approach focused on a metastable neutralization-sensitive site called antigenic site Ø (zero) at the membrane-distal apex of the RSV fusion (F) glycoprotein. Immunization of mice and nonhuman primates with a site Ø-stabilized version of RSV F (called DS-Cav1) elicited antibodies many times the protective threshold (McLellan 2013 Science 342, 592-598) with clinical trials indicating the prefusion-stabilized immunogen to substantially boost RSV-neutralizing titers in healthy adults (Crank 2019 365, 505-509).
Biography
Education
Ph.D., Chinese Academy of Sciences, Beijing, China
M.Sc., Wayne State University, Detroit, MI
Dr. Tongqing Zhou joined the Dale and Betty Bumpers Vaccine Research Center in 2001, became a staff scientist in 2005, and was promoted to chief of the NIAID Structural Bioinformatics Core in 2018. Dr. Zhou received his Ph.D. in cell biology from the Chinese Academy of Sciences and his M.Sc. in electronic and computer controlled systems from Wayne State University. He is joined by the two co-heads of the core, Drs. Gwo-Yu Chuang and Chen-Hsiang Shen. Dr. Gwo-Yu Chuang received his Ph.D. training under the mentorship of Dr. Sandor Vajda, Boston University. Dr. Shen received his Ph.D. training under the mentorship of Dr. Irene Weber, Georgia State University. Drs. Peter Kwong and Lawrence Shapiro also serve as advisory mentors to the Structural Virology and Vaccinology Section.
Selected Publications
Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, Louder MK, O'Dell S, Rawi R, Sastry M, Shen CH, Zhang B, Zhou T, Asokan M, Bailer RT, Chambers M, Chen X, Choi CW, Dandey VP, Doria-Rose NA, Druz A, Eng ET, Farney SK, Foulds KE, Geng H, Georgiev IS, Gorman J, Hill KR, Jafari AJ, Kwon YD, Lai YT, Lemmin T, McKee K, Ohr TY, Ou L, Peng D, Rowshan AP, Sheng Z, Todd JP, Tsybovsky Y, Viox EG, Wang Y, Wei H, Yang Y, Zhou AF, Chen R, Yang L, Scorpio DG, McDermott AB, Shapiro L, Carragher B, Potter CS, Mascola JR, Kwong PD. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med. 2018 Jun;24(6): 857-867.
Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016 May 5;165(4):813-26. doi: 10.1016/j.cell.2016.04.010.
Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014 Oct 23;514(7523):455-61.
McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013 Nov 1;342(6158):592-8.
Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010 Aug 13;329(5993):811-7.
Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998 Jun 18;393(6686):648-59.
Structural Models
Visit the RCSB PDB for a listing of SVVS structures.

