DNA Tumor Virus Section
Established in 1994
Alison McBride, Ph.D.
Chief, DNA Tumor Virus Section

Major Areas of Research
- Mechanisms by which extrachromosomal human papillomavirus (HPV) genomes are established, partitioned, and amplified during persistent infection.
- The role of host intrinsic immunity, and DNA damage response and repair pathways in HPV DNA replication
- Analysis of the mechanism and consequences of viral genome integration in HPV-associated cancers
- Keratinocyte biology: Reprogramming Keratinocytes by Rho Kinase inhibition
- PaVE: The PapillomaVirus Episteme, a bioinformatics resource
Program Description
Papillomaviruses persistently infect and replicate in stratified cutaneous and mucosal epithelia. In most cases, the infections are benign and self-limiting, but persistent infection with certain papillomavirus types can lead to the development of carcinomas. In fact, HPV is the cause of about 5% of human cancers.
HPVs infect the basal cells of stratified epithelia and the viral DNA replicates in these dividing cells as low copy, extrachromosomal circular DNA molecules. Viral genome amplification and synthesis of capsid proteins occurs only in the upper layers of the epithelium. Papillomavirus infections are usually long-lived, and the basal cells provide a reservoir of infected cells for the overlying virus producing tissue. This strategy requires that the papillomaviruses have a faithful and robust mechanism to replicate and retain their extrachromosomal genomes in the nuclei of dividing cells. We study all aspects of HPV replication using primary cell culture, genomics, transcriptomics, proteomics, and advanced imaging techniques.
Gallery of our work:
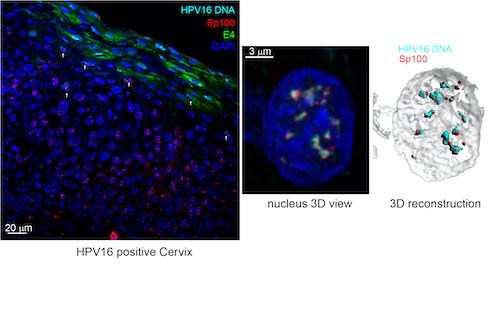
Association of the Sp100 protein with viral replication foci in a cervical lesion. Created by James Stamos and Alison McBride.

The PapillomaVirus Episteme: A Sequence Database

HPV DNA is often integrated in cancers. Here, tandemly integrated HPV genomes are identified by Fiber-FISH. Cellular DNA is stained in red and viral DNA in green. Created by Catherine Redmond and Alison McBride.
Videos
HPVs will only replicate in host keratinocytes. We discovered that primary keratinocytes could be sustained in a conditionally proliferative state by culture in the presence of Rho kinase inhibitors. This discovery (Patent US8637310) has revolutionized the procurement and culture of primary cells. Video created by Alison McBride.
Credit: NIAID
Biography
Education
B.Sc., University of Glasgow, Scotland, UK
Ph.D., Imperial Cancer Research Fund and Imperial College, UK
Dr. McBride received a B.Sc. (Hons) in Molecular Biology from the University of Glasgow, Scotland, and a Ph.D. in Biochemistry from the Imperial Cancer Research Fund and Imperial College, England, studying Epstein-Barr virus. She began working on human and other papillomaviruses as a postdoctoral fellow in the National Cancer Institute and joined NIAID in 1994. She became a senior investigator in the Laboratory of Viral Diseases in 2000, and a section chief in 2001. Dr. McBride is also adjunct faculty, a member of the Virology Graduate Program at the University of Maryland. She is a fellow, American Academy of Microbiology; Section Editor, PLOS Pathogens; and Editor, Virology.
Selected Publications
Warburton A, Markowitz TE, Katz JP, Pipas JM, McBride AA. Recurrent integration of human papillomavirus genomes at transcriptional regulatory hubs. NPJ Genom Med. 2021 Nov 30;6(1):101.
Khurana S, Markowitz TE, Kabat J, McBride AA. Spatial and Functional Organization of Human Papillomavirus Replication Foci in the Productive Stage of Infection. mBio. 2021 Dec 21;12(6):e0268421.
McBride AA. Human papillomaviruses: diversity, infection and host interactions. Nat Rev Microbiol. 2022 Feb;20(2):95-108.
Coursey TL, Van Doorslaer K, McBride AA. Regulation of Human Papillomavirus 18 Genome Replication, Establishment, and Persistence by Sequences in the Viral Upstream Regulatory Region. J Virol. 2021 Sep 9;95(19):e0068621.
Porter SS, Liddle JC, Browne K, Pastrana DV, Garcia BA, Buck CB, Weitzman MD, McBride AA. Histone Modifications in Papillomavirus Virion Minichromosomes. mBio. 2021 Feb 16;12(1):e03274-20.
Stepp WH, Stamos JD, Khurana S, Warburton A, McBride AA. Sp100 colocalizes with HPV replication foci and restricts the productive stage of the infectious cycle. PLoS Pathog. 2017 Oct 2;13(10):e1006660.
Active Projects and Available Positions
The McBride lab is recruiting post-baccalaureate and post-doctoral trainees. Interested candidates should send their CV, a one-page letter of interest and professional references to LVDResearchOpport@niaid.nih.gov or directly to Dr. McBride.
Ongoing projects:
Project 1: Studying how HPVs hijack nuclear processes for viral replication and transcription using spatial/in situ imaging techniques
Project 2: Defining mechanisms by which papillomavirus genomes are established, partitioned, and amplified in keratinocytes during persistent infection
Project 3: Defining innate immune factors that modulate HPV DNA replication
Project 4: Designing anti-viral therapeutics to resolve persistent HPV infection
Project 5: Studying how HPV replication mechanisms can promote accidental integration and HPV-associated cancers
Mentorship
Dr. McBride is Adjunct Faculty at the University of Maryland and serves as a mentor for the Virology Program.
Research Group
The DTVS studies mechanisms and strategies of HPV replication.


