The Twinbrook Imaging Facility’s mission is to provide cutting edge instrumentation, instruction, and technical support to enable researchers to acquire and analyze high-quality images of living and fixed cells from all kinds of model systems. Support is available and encouraged through all phases of a research question, including study design, data acquisition, analysis, and publication. In addition, the facility strives to train researchers to become excellent microscopists in their own right. Active communication amongst users is cultivated to culture academic growth and solve common problems.
Imaging Systems
The facility has experience with and supports a number of light microscopy techniques to visualize live and fixed cells. All imaging techniques are limited by tradeoffs in Speed, Resolution, Phototoxicity (Gentleness), and Depth, but the comprehensive instrumentation available at the facility provides excellent outcomes in each parameter.
Explore the chart below or access an accessible table to discover the instrumentation available at the facility.
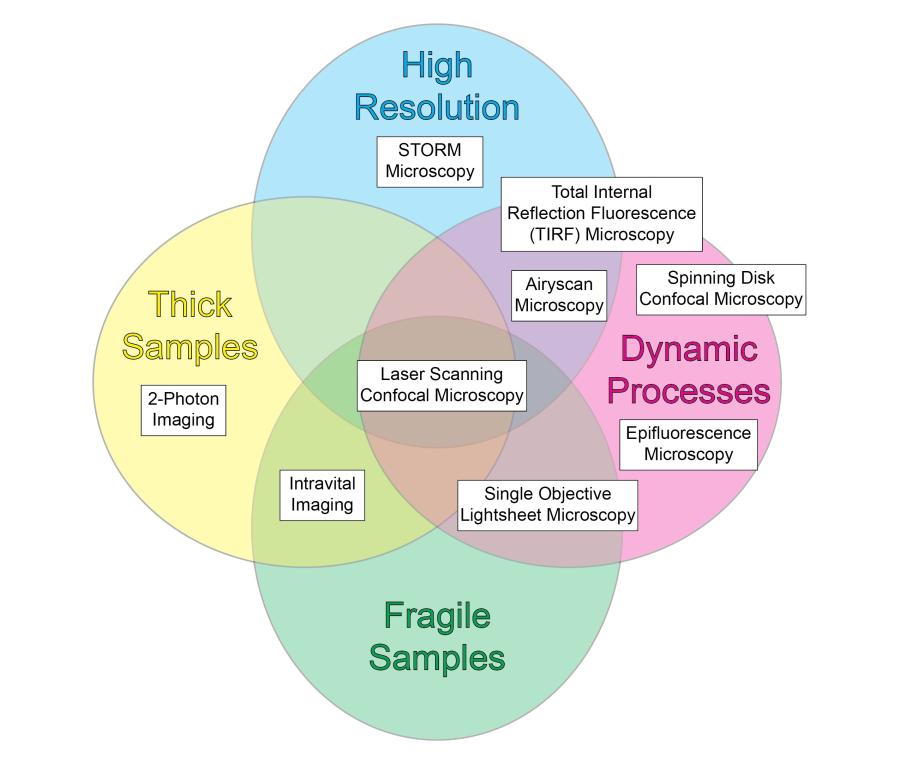
Additionally, we offer analytical techniques such as Forster Resonance Transfer (FRET), fluorescence lifetime imaging (FLIM), and fluorescence correlation spectroscopy (FCS). Overall, the Facility strives to maintain fluidity in the open design of its microscopes to meet the changing needs of the investigators. Contact Dr. Brzostowski for further information or to schedule time on one of the instruments.
Data Analysis Services
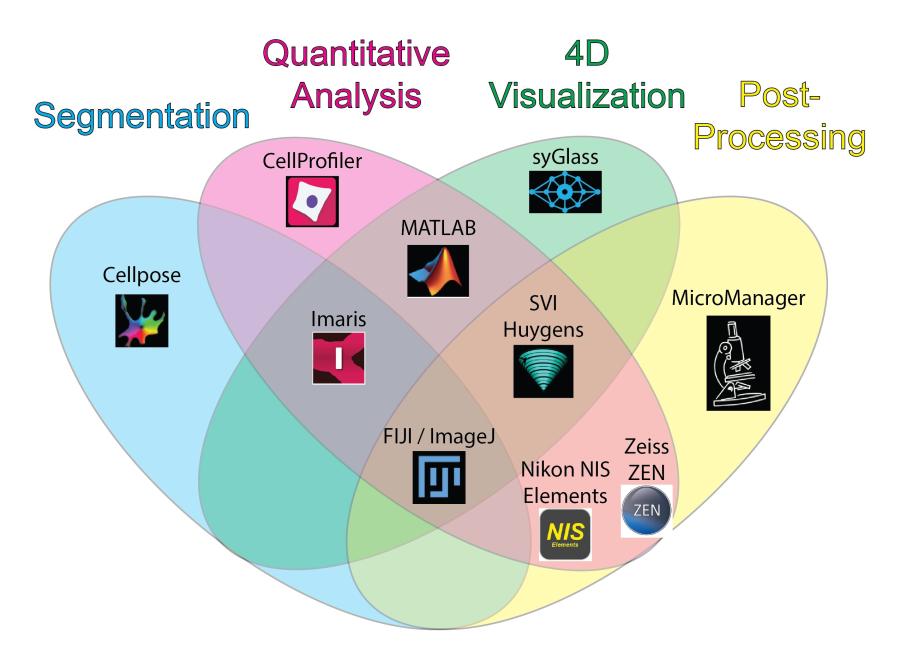
In addition to the instrumentation provided, the Twinbrook Facility offers custom imaging data analysis services. Users have access to unlimited data storage with automatic backup functionality. Access to a suite of cutting-edge open source and proprietary image visualization and analysis software packages on two high end computing workstations is also provided (see below chart). Dr. Maria Traver develops and implements custom image analysis pipelines and is available for consultations and training in quantitative analysis techniques. Contact Dr. Traver for more information or to schedule a time to discuss services.
Explore the chart below or access an accessible table to discover the major strengths of each software package available at the facility.
Selected Publications
The Twinbrook Imaging Facility and its users strive to share their knowledge and have published numerous book chapters, methods papers, and journal articles.
Dacon C, Moskovitz R, Swearingen K, Da Silva Pereira L, Flores-Garcia Y, Aleshnick M, Kanatani S, Flynn B, Molina-Cruz A, Wollenberg K, Traver M, Kirtley P, Purser L, Dillon M, Bonilla B, Franco A, Petros S, Kritzberg J, Tucker C, Paez GG, Gupta P, Shears MJ, Pazzi J, Edgar JM, Teng AA, Belmonte A, Oda K, Doumbo S, Krymskaya L, Skinner J, Li S, Ghosal S, Kayentao K, Ongoiba A, Vaughan A, Campo JJ, Traore B, Barillas-Mury C, Wijayalath W, Idris A, Crompton PD, Sinnis P, Wilder BK, Zavala F, Seder RA, Wilson IA, Tan J. Protective antibodies target cryptic epitope unmasked by cleavage of malaria sporozoite protein. Science. 2025 Jan 3;387(6729):eadr0510.
Xu X, Ha H, Brzostowski J, Jin T. Quantitative Monitoring of GPCR-Mediated Spatiotemporal IP3 Dynamics Using Confocal Fluorescence Microscopy. Methods Mol Biol. 2024;2814:195-207.
Yang RS, Traver M, Barefoot N, Stephens T, Alabanza C, Manzella-Lapeira J, Zou G, Wolff J, Li Y, Resto M, Shadrick W, Yang Y, Ivleva VB, Tsybovsky Y, Carlton K, Brzostowski J, Gall JG, Lei QP. Mosaic quadrivalent influenza vaccine single nanoparticle characterization. Sci Rep. 2024 Feb 24;14(1):4534.
Brzostowski J, Sohn HW eds. Methods in Molecular Biology: Confocal Microscopy. Springer, 2021.
Brzostowski J. General Considerations for Acquiring a Three-Color Image by Laser Scanning Confocal Microscopy. Methods Mol Biol. 2021;2304:65-91.
Manzella-Lapeira J, Brzostowski J. Fluorescence Lifetime Imaging as a Noninvasive Tool to Study Plasmodium Falciparum Metabolism. Methods Mol Biol. 2021;2304:301-313.
Manzella-Lapeira J, Brzostowski J, Serra-Vinardell J. Studying Neuronal Biology Using Spinning Disc Confocal Microscopy. Methods Mol Biol. 2021;2304:265-283.
Manzella-Lapeira J, Brzostowski JA. Imaging Protein-Protein Interactions by Förster Resonance Energy Transfer (FRET) Microscopy in Live Cells. Curr Protoc Protein Sci. 2018 Aug;93(1):e58.
Sohn HW, Brzostowski J. Time-Lapse Förster Resonance Energy Transfer Imaging by Confocal Laser Scanning Microscopy for Analyzing Dynamic Molecular Interactions in the Plasma Membrane of B Cells. Methods Mol Biol. 2018;1707:207-224.

